Curriculum Vitae – Robin L. McCarley - LSU Department of ...
Curriculum Vitae – Robin L. McCarley - LSU Department of ...
Curriculum Vitae – Robin L. McCarley - LSU Department of ...
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
<strong>Robin</strong> L. <strong>McCarley</strong> September 2011<br />
For all <strong>of</strong> the stimuli-responsive s<strong>of</strong>t matter systems, we foresee numerous target stimuli as possible<br />
avenues <strong>of</strong> investigation, such as amidases, esterases, and small-molecule biomarkers. Thus, the variety<br />
<strong>of</strong> possible materials we can make will have a significant impact on numerous application areas in<br />
nanomedicine, both from diagnostic and possible treatment avenues.<br />
Increasing the Functional Capabilities <strong>of</strong> Micr<strong>of</strong>luidic Devices by Surface Chemical<br />
Modification and Creation <strong>of</strong> Nanostructures within Microstructures (NIH and NSF funding)<br />
Overview. Our group is a pioneer in the chemical modification <strong>of</strong> polymer surfaces that allow for<br />
attachment <strong>of</strong> key biological species, small molecules, and metallic features to the polymer surface, so as<br />
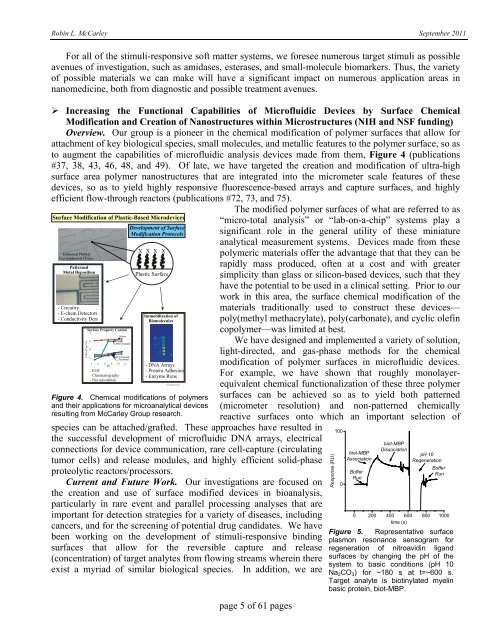
to augment the capabilities <strong>of</strong> micr<strong>of</strong>luidic analysis devices made from them, Figure 4 (publications<br />
#37, 38, 43, 46, 48, and 49). Of late, we have targeted the creation and modification <strong>of</strong> ultra-high<br />
surface area polymer nanostructures that are integrated into the micrometer scale features <strong>of</strong> these<br />
devices, so as to yield highly responsive fluorescence-based arrays and capture surfaces, and highly<br />
efficient flow-through reactors (publications #72, 73, and 75).<br />
Surface Modification <strong>of</strong> Plastic-Based Microdevices<br />
Development <strong>of</strong> Surface<br />
Modification Protocols<br />
Embossed PMMA<br />
Electrophoresis Device<br />
Patterned<br />
Metal Deposition<br />
- Circuitry<br />
- E-chem Detectors<br />
- Conductivity Dets<br />
EOF(x10 -4 cm 2 EOF(x10 /Vs)<br />
-4 cm 2 /Vs)<br />
4<br />
2<br />
0<br />
Pristine<br />
COOH-Terminated<br />
-2<br />
NH -Terminated<br />
2<br />
C -Terminated<br />
18<br />
2 4 6 8<br />
pH<br />
10 12<br />
X X X X<br />
Plastic Surface<br />
Immobilization <strong>of</strong><br />
Biomolecules<br />
- DNA Arrays<br />
- Protein Adhesion<br />
- Enzyme Rxns<br />
ChemModMachOver<br />
Figure 4. Chemical modifications <strong>of</strong> polymers<br />
and their applications for microanalytical devices<br />
resulting from <strong>McCarley</strong> Group research.<br />
species can be attached/grafted. These approaches have resulted in<br />
the successful development <strong>of</strong> micr<strong>of</strong>luidic DNA arrays, electrical<br />
connections for device communication, rare cell-capture (circulating<br />
tumor cells) and release modules, and highly efficient solid-phase<br />
proteolytic reactors/processors.<br />
Current and Future Work. Our investigations are focused on<br />
the creation and use <strong>of</strong> surface modified devices in bioanalysis,<br />
particularly in rare event and parallel processing analyses that are<br />
important for detection strategies for a variety <strong>of</strong> diseases, including<br />
cancers, and for the screening <strong>of</strong> potential drug candidates. We have<br />
been working on the development <strong>of</strong> stimuli-responsive binding<br />
surfaces that allow for the reversible capture and release<br />
(concentration) <strong>of</strong> target analytes from flowing streams wherein there<br />
exist a myriad <strong>of</strong> similar biological species. In addition, we are<br />
The modified polymer surfaces <strong>of</strong> what are referred to as<br />
“micro-total analysis” or “lab-on-a-chip” systems play a<br />
significant role in the general utility <strong>of</strong> these miniature<br />
analytical measurement systems. Devices made from these<br />
polymeric materials <strong>of</strong>fer the advantage that that they can be<br />
rapidly mass produced, <strong>of</strong>ten at a cost and with greater<br />
simplicity than glass or silicon-based devices, such that they<br />
have the potential to be used in a clinical setting. Prior to our<br />
work in this area, the surface chemical modification <strong>of</strong> the<br />
materials traditionally used to construct these devices—<br />
poly(methyl methacrylate), poly(carbonate), and cyclic olefin<br />
copolymer—was limited at best.<br />
We have designed and implemented a variety <strong>of</strong> solution,<br />
light-directed, and gas-phase methods for the chemical<br />
modification <strong>of</strong> polymer surfaces in micr<strong>of</strong>luidic devices.<br />
For example, we have shown that roughly monolayerequivalent<br />
chemical functionalization <strong>of</strong> these three polymer<br />
surfaces can be achieved so as to yield both patterned<br />
(micrometer resolution) and non-patterned chemically<br />
reactive surfaces onto which an important selection <strong>of</strong><br />
page 5 <strong>of</strong> 61 pages<br />
Response (RU)<br />
100<br />
0<br />
biot-MBP<br />
Association<br />
Buffer<br />
Run<br />
biot-MBP<br />
Dissociation<br />
pH 10<br />
Regeneration<br />
Buffer<br />
Run<br />
0 200 400 600 800 1000<br />
time (s)<br />
Figure 5. Representative surface<br />
plasmon resonance sensogram for<br />
regeneration <strong>of</strong> nitroavidin ligand<br />
surfaces by changing the pH <strong>of</strong> the<br />
system to basic conditions (pH 10<br />
Na 2CO 3) for ~180 s at t=~600 s.<br />
Target analyte is biotinylated myelin<br />
basic protein, biot-MBP.