Algorithms on Sequences
Algorithms on Sequences
Algorithms on Sequences
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
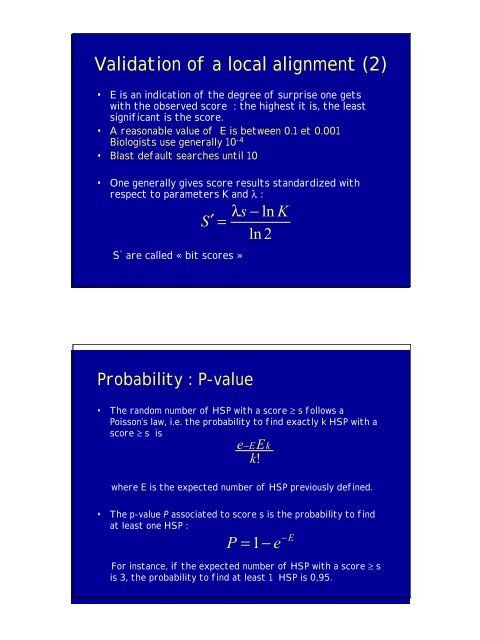
Validati<strong>on</strong> of a local alignment (2)<br />
• E is an indicati<strong>on</strong> of the degree of surprise <strong>on</strong>e gets<br />
with the observed score : the highest it is, the least<br />
significant is the score.<br />
• A reas<strong>on</strong>able value of E is between 0.1 et 0.001<br />
Biologists use generally 10 -4<br />
• Blast default searches until 10<br />
• One generally gives score results standardized with<br />
respect to parameters K and λ :<br />
S ' are called « bit scores »<br />
s − ln K<br />
S′<br />
=<br />
λ<br />
Probability : P-value<br />
ln 2<br />
• The random number of HSP with a score ≥ s follows a<br />
Poiss<strong>on</strong>’s law, i.e. the probability to find exactly k HSP with a<br />
score ≥ s is<br />
e k E −<br />
where E is the expected number of HSP previously defined.<br />
• The p-value P associated to score s is the probability to find<br />
at least <strong>on</strong>e HSP :<br />
−E<br />
P<br />
k! E<br />
= 1−<br />
For instance, if the expected number of HSP with a score ≥ s<br />
is 3, the probability to find at least 1 HSP is 0,95.<br />
e