Molecular characterisation of SGCE-associated myoclonus-dystonia ...
Molecular characterisation of SGCE-associated myoclonus-dystonia ...
Molecular characterisation of SGCE-associated myoclonus-dystonia ...
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
Patients, material and methods 33<br />
PCR reaction according to the manufacturer’s protocol (Fig 5). Amplification products were<br />
separated by polyacrylamide gel electrophoresis (PAGE, see 2.3.3.2) and visualized on an<br />
automated sequencing machine. For all samples, a specific pattern <strong>of</strong> bands, varying in size<br />
and intensity, was observed. Each <strong>of</strong> these bands represented one amplified pair <strong>of</strong> probes<br />
from the kit. The MLPA gel images were analyzed with the help <strong>of</strong> an image processing<br />
program (TotalLab), employing a “nearest neighbour” model (Djarmati et al., 2007), which<br />
allows calculating the relative initial concentration <strong>of</strong> the specific DNA fragment. Normalized<br />
value ratios between 0.8 and 1.2 were considered normal, a heterozygous deletion was<br />
expected at a ratio between 0.3 and 0.7, and a heterozygous duplication between 1.3 and 1.7.<br />
MLPA was performed for all exons <strong>of</strong> <strong>SGCE</strong> except the rare splicing variant, exon 10, in the<br />
M-D approach (Fig 9).<br />
Hybridisation sequence A’ and A’’<br />
Primer X<br />
Stuffer sequence A<br />
Primer Y<br />
Hybridisation sequence B’ and B’’<br />
Primer X<br />
Primer Y<br />
Stuffer sequence B<br />
1. Hybridisation<br />
3’ 5’<br />
Target A<br />
3’ 5’<br />
Target B<br />
2. Ligation<br />
3’ 5’<br />
Target A<br />
3’ 5’<br />
Target B<br />
3. Amplification<br />
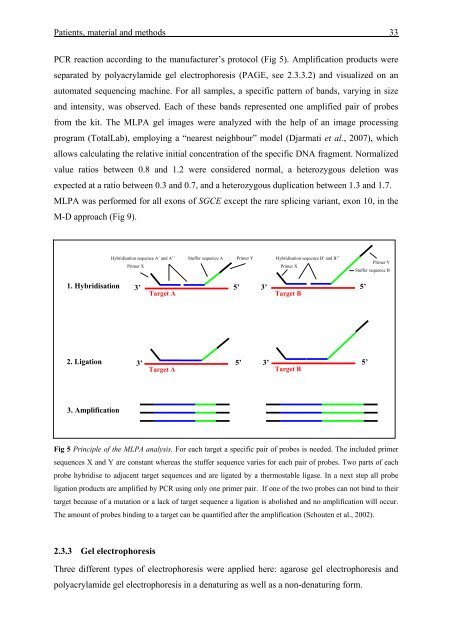
Fig 5 Principle <strong>of</strong> the MLPA analysis. For each target a specific pair <strong>of</strong> probes is needed. The included primer<br />
sequences X and Y are constant whereas the stuffer sequence varies for each pair <strong>of</strong> probes. Two parts <strong>of</strong> each<br />
probe hybridise to adjacent target sequences and are ligated by a thermostable ligase. In a next step all probe<br />
ligation products are amplified by PCR using only one primer pair. If one <strong>of</strong> the two probes can not bind to their<br />
target because <strong>of</strong> a mutation or a lack <strong>of</strong> target sequence a ligation is abolished and no amplification will occur.<br />
The amount <strong>of</strong> probes binding to a target can be quantified after the amplification (Schouten et al., 2002).<br />
2.3.3 Gel electrophoresis<br />
Three different types <strong>of</strong> electrophoresis were applied here: agarose gel electrophoresis and<br />
polyacrylamide gel electrophoresis in a denaturing as well as a non-denaturing form.