Bacterial Identification Single Pathogen Identification by 16S rRNA ...
Bacterial Identification Single Pathogen Identification by 16S rRNA ...
Bacterial Identification Single Pathogen Identification by 16S rRNA ...
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
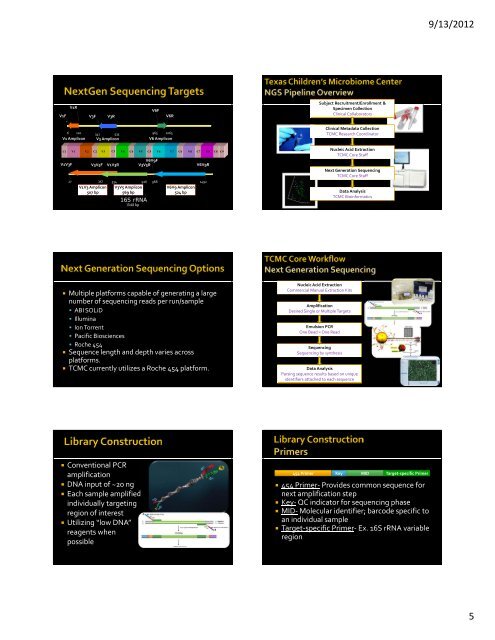
9/13/2012V1FV1RV3FV3RV6FV6RSubject Recruitment/Enrollment &Specimen CollectionClinical Collaborators6 120 341 533965 1065V1 Amplicon V3 AmpliconV6 AmpliconV1 V2 V3 V4 V5 V6 V7 V8 V9C1 C2 C3 C4 C5 C6 C7 C8 C9V1V3FV3V5FV1V3 Amplicon507 bpV1V3R<strong>16S</strong> <strong>rRNA</strong>1542 bpV6V9FV3V5RV6V9R27 357 534 926 968 1492V3V5 Amplicon569 bpV6V9 Amplicon524 bpClinical Metadata CollectionTCMC Research CoordinatorNucleic Acid ExtractionTCMC Core StaffNext Generation SequencingTCMC Core StaffData AnalysisTCMC Bioinformatics• Multiple platforms capable of generating a largenumber of sequencing reads per run/sample• ABI SOLiD• Illumina• Ion Torrent• Pacific Biosciences• Roche 454• Sequence length and depth varies acrossplatforms.• TCMC currently utilizes a Roche 454 platform.Nucleic Acid ExtractionCommercial Manual Extraction KitsAmplificationDesired <strong>Single</strong> or Multiple TargetsEmulsion PCROne Bead = One ReadSequencingSequencing <strong>by</strong> synthesisData AnalysisParsing sequence results based on uniqueidentifiers attached to each sequence• Conventional PCRamplification• DNA input of ~20 ng• Each sample amplifiedindividually targetingregion of interest• Utilizing “low DNA”reagents whenpossible454 Primer Key MID Target‐specific Primer• 454 Primer‐ Provides common sequence fornext amplification step• Key‐ QC indicator for sequencing phase• MID‐ Molecular identifier; barcode specific toan individual sample• Target‐specific Primer‐ Ex. <strong>16S</strong> <strong>rRNA</strong> variableregion5