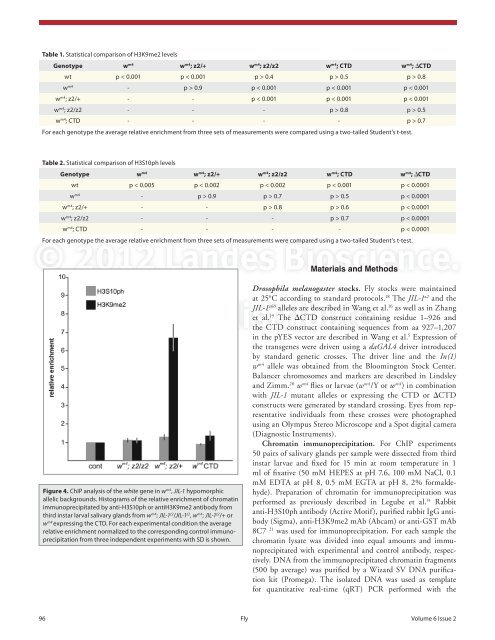
Table 1. Statistical comparison of H3K9me2 levelsGenotype w m4 w m4 ; z2/+ w m4 ; z2/z2 w m4 ; CTD w m4 ; ΔCTDwt p < 0.001 p < 0.001 p > 0.4 p > 0.5 p > 0.8w m4 - p > 0.9 p < 0.001 p < 0.001 p < 0.001w m4 ; z2/+ - - p < 0.001 p < 0.001 p < 0.001w m4 ; z2/z2 - - - p > 0.8 p > 0.5w m4 ; CTD - - - - p > 0.7For each genotype the average relative enrichment from three sets of measurements were compared using a two-tailed Student’s t-test.Table 2. Statistical comparison of H3S10ph levelsGenotype w m4 w m4 ; z2/+ w m4 ; z2/z2 w m4 ; CTD w m4 ; ΔCTDwt p < 0.005 p < 0.002 p < 0.002 p < 0.001 p < 0.0001w m4 - p > 0.9 p > 0.7 p > 0.5 p < 0.0001w m4 ; z2/+ - - p > 0.8 p > 0.6 p < 0.0001w m4 ; z2/z2 - - - p > 0.7 p < 0.0001w m4 ; CTD - - - - p < 0.0001For each genotype the average relative enrichment from three sets of measurements were compared using a two-tailed Student’s t-test.© 2012 <strong>Landes</strong> <strong>Bioscience</strong>.Materials and MethodsDrosophila melanogaster stocks. Fly stocks were maintainedat 25°C according to standard protocols. 18 The JIL-1 z2 and theJIL-1 z60 alleles are described in Wang et al. 16 as well as in Zhanget al. 19 The ΔCTD construct containing residue 1–926 andDo not distribute.the CTD construct containing sequences from aa 927–1,207Figure 4. ChIP analysis of the white gene in w m4 , JIL-1 hypomorphicallelic backgrounds. Histograms of the relative enrichment of chromatinimmunoprecipitated by anti-H3S10ph or antiH3K9me2 antibody fromthird instar larval salivary glands from w m4 ; JIL-1 z2 /JIL-1 z2 , w m4 ; JIL-1 z2 /+ orw m4 expressing the CTD. For each experimental condition the averagerelative enrichment normalized to the corresponding control immunoprecipitationfrom three independent experiments with SD is shown.in the pYES vector are described in Wang et al. 5 Expression ofthe transgenes were driven using a daGAL4 driver introducedby standard genetic crosses. The driver line and the In(1)w m4 allele was obtained from the Bloomington Stock Center.Balancer chromosomes and markers are described in Lindsleyand Zimm. 20 w m4 flies or larvae (w m4 /Y or w m4 ) in combinationwith JIL-1 mutant alleles or expressing the CTD or ΔCTDconstructs were generated by standard crossing. Eyes from representativeindividuals from these crosses were photographedusing an Olympus Stereo Microscope and a Spot digital camera(Diagnostic Instruments).Chromatin immunoprecipitation. For ChIP experiments50 pairs of salivary glands per sample were dissected from thirdinstar larvae and fixed for 15 min at room temperature in 1ml of fixative (50 mM HEPES at pH 7.6, 100 mM NaCl, 0.1mM EDTA at pH 8, 0.5 mM EGTA at pH 8, 2% formaldehyde).Preparation of chromatin for immunoprecipitation wasperformed as previously described in Legube et al. 14 Rabbitanti-H3S10ph antibody (Active Motif), purified rabbit IgG antibody(Sigma), anti-H3K9me2 mAb (Abcam) or anti-GST mAb8C7 21 was used for immunoprecipitation. For each sample thechromatin lysate was divided into equal amounts and immunoprecipitatedwith experimental and control antibody, respectively.DNA from the immunoprecipitated chromatin fragments(500 bp average) was purified by a Wizard SV DNA purificationkit (Promega). The isolated DNA was used as templatefor quantitative real-time (qRT) PCR performed with the96 Fly Volume 6 Issue 2
Stratagene Mx4000 real-time cycler. The PCR mixture containedBrilliant II SYBR Green QPCR Master Mix (Stratagene) as wellas the corresponding primers: white-forward 5'-GTG CTG TGCCAA AAC TCC TC-3', white-reverse 5'-GAT GCT CGG CAGATG GGT TGT-3' which amplify region -110 to +65. Cyclingparameters were 10 min at 95°C, followed by 40 cycles of 30 secat 95°C, 30 sec at 55°C and 30 sec at 72°C. Fluorescence intensitieswere plotted against the number of cycles using an algorithmprovided by Stratagene. DNA levels were quantified using a calibrationcurve based on dilution of concentrated DNA. For eachexperimental condition the relative enrichment was normalizedto the corresponding control immunoprecipitation from thesame chromatin lysate.Disclosure of Potential Conflicts of InterestNo potential conflicts of interest were disclosed.AcknowledgmentsWe thank members of the laboratory for discussion, advice andcritical reading of the manuscript. We also wish to acknowledgeMr. Atrez Norwood for technical assistance. This work was supportedby NIH Grant GM062916 (K.M.J./J.J.).References1. Zhang W, Deng H, Bao X, Lerach S, Girton J,Johansen J, et al. The JIL-1 histone H3S10 kinaseregulates dimethyl H3K9 modifications and heterochromaticspreading in Drosophila. Development2006; 133:229-35; PMID:16339185; http://dx.doi.org/10.1242/dev.02199.2. Deng H, Bao X, Zhang W, Girton J, Johansen J,Johansen KM. Reduced levels of Su(var)3–9 but notSu(var)2–5 (HP1) counteract the effects on chromatinstructure and viability in loss-of-functionmutants of the JIL-1 histone H3S10 kinase. Genetics2007; 177:79-87; PMID:17660558; http://dx.doi.org/10.1534/genetics.107.075143.3. Deng H, Cai W, Wang C, Lerach S, Delattre M, Girton7. Bao X, Deng H, Johansen J, Girton J, Johansen KM.Loss-of-function alleles of the JIL-1 histone H3S10kinase enhance position-effect variegation at pericentricsites in Drosophila heterochromatin. Genetics2007; 176:1355-8; PMID:17435241; http://dx.doi.org/10.1534/genetics.107.073676.8. Wang C, Girton J, Johansen J, Johansen KM. Abalance between euchromatic (JIL-1) and heterochromatic[SU(var)2–5 and SU(var)3–9] factors regulatesposition-effect variegation in Drosophila. Genetics2011; 188:745-8; PMID:21515582; http://dx.doi.org/10.1534/genetics.111.129353.9. Muller HJ. Types of visible variegations induced byX-rays in Drosophila. J Genet 1930; 22:299-335;http://dx.doi.org/10.1007/BF02984195.15. Bao X, Cai W, Deng H, Zhang W, Krencik R, GirtonJ, et al. The COOH-terminal domain of the JIL-1histone H3S10 kinase interacts with histone H3 andis required for correct targeting to chromatin. J BiolChem 2008; 283:32741-50; PMID:18819909; http://dx.doi.org/10.1074/jbc.M806227200.16. Wang Y, Zhang W, Jin Y, Johansen J, Johansen KM.The JIL-1 tandem kinase mediates histone H3 phosphorylationand is required for maintenance of chromatinstructure in Drosophila. Cell 2001; 105:433-43;PMID:11371341; http://dx.doi.org/10.1016/S0092-8674(01)00325-7.17. Cai W, Bao X, Deng H, Jin Y, Girton J, Johansen J, etal. RNA polymerase II-mediated transcription at active© 2012 <strong>Landes</strong> <strong>Bioscience</strong>.loci does not require histone H3S10 phosphoryla-J, et al. JIL-1 and Su(var)3–7 interact genetically andcounteract each other’s effect on position-effect variegationin Drosophila. Genetics 2010; 185:1183-92;PMID:20457875; http://dx.doi.org/10.1534/genetics.110.117150.4. Ebert A, Schotta G, Lein S, Kubicek S, Krauss V,Jenuwein T, et al. Su(var) genes regulate the balancebetween euchromatin and heterochromatinin Drosophila. Genes Dev 2004; 18:2973-83;PMID:15574598; http://dx.doi.org/10.1101/gad.323004.5. Wang C, Cai W, Li Y, Deng H, Bao X, Girton J, etal. The epigenetic H3S10 phosphorylation mark isrequired for counteracting heterochromatic spreadingand gene silencing in Drosophila melanogaster. J Cell Sci2011; 124:4309-17; PMID:22247192; http://dx.doi.org/10.1242/jcs.092585.6. Lerach S, Zhang W, Bao X, Deng H, Girton J,Johansen J, et al. Loss-of-function alleles of the JIL-1kinase are strong suppressors of position effect variegationof the w m4 allele in Drosophila. Genetics2006; 173:2403-6; PMID:16702418; http://dx.doi.org/10.1534/genetics.106.059253.10. Pirrotta V, Hadfield C, Pretorius GHJ. Microdissectionand cloning of the white locus and the 3B1-3C2 regionof the Drosophila X chromosome. EMBO J 1983;2:927-34; PMID:16453459.11. Grewal SI, Elgin SC. Heterochromatin: new possibilitiesfor the inheritance of structure. Curr OpinGenet Dev 2002; 12:178-87; PMID:11893491; http://dx.doi.org/10.1016/S0959-437X(02)00284-8.12. Weiler KS, Wakimoto BT. Heterochromatin andtion in Drosophila. Development 2008; 135:2917-25; PMID:18667461; http://dx.doi.org/10.1242/dev.024927.18. Roberts DB. In Drosophila: A Practical Approach IRLPress, Oxford UK 1998.19. Zhang W, Jin Y, Ji Y, Girton J, Johansen J, JohansenKM. Genetic and phenotypic analysis of alleles of theDrosophila chromosomal JIL-1 kinase reveals a func-Do not distribute.tional requirement at multiple developmental stages.gene expression in Drosophila. Annu Rev Genet1995; 29:577-605; PMID:8825487; http://dx.doi.org/10.1146/annurev.ge.29.120195.003045.13. Girton JR, Johansen KM. Chromatin structureand the regulation of gene expression: the lessonsof PEV in Drosophila. Adv Genet 2008; 61:1-43;PMID:18282501; http://dx.doi.org/10.1016/S0065-2660(07)00001-6.14. Legube G, McWeeney SK, Lercher MJ, Akhtar A.X-chromosome-wide profiling of MSL-1 distributionand dosage compensation in Drosophila. Genes Dev2006; 20:871-83; PMID:16547175; http://dx.doi.org/10.1101/gad.377506.Genetics 2003; 165:1341-54; PMID:14668387.20. Lindsley DL, Zimm GG. The genome of Drosophilamelanogaster. Academic Press, New York NY 1992.21. Rath U, Wang D, Ding Y, Xu YZ, Qi H, Blacketer MJ,et al. Chromator, a novel and essential chromodomainprotein interacts directly with the putative spindlematrix protein skeletor. J Cell Biochem 2004; 93:1033-47; PMID:15389869; http://dx.doi.org/10.1002/jcb.20243.www.landesbioscience.com Fly 97