pTriEx-4 Neo Vector - Gene Synthesis
pTriEx-4 Neo Vector - Gene Synthesis
pTriEx-4 Neo Vector - Gene Synthesis
- No tags were found...
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
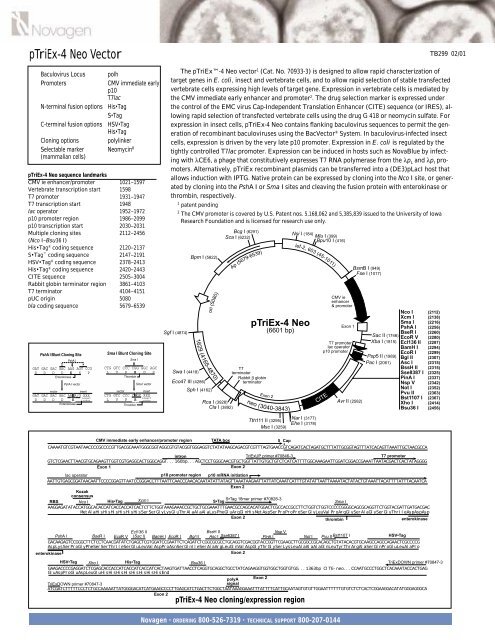
<strong>pTriEx</strong>-4 <strong>Neo</strong> <strong>Vector</strong>Baculovirus Locus polhPromotersCMV immediate earlyp10T7lacN-terminal fusion options His•TagS•TagC-terminal fusion options HSV•TagHis•TagCloning options polylinkerSelectable marker <strong>Neo</strong>mycin R(mammalian cells)<strong>pTriEx</strong>-4 <strong>Neo</strong> sequence landmarksCMV ie enhancer/promoter 1021–1597Vertebrate transcription start 1598T7 promoter 1931–1947T7 transcription start 1948lac operator 1952–1972p10 promoter region 1986–2099p10 transcription start 2030–2031Multiple cloning sites 2112–2456(Nco I–Bsu36 I)His•Tag ® coding sequence 2120–2137S•Tag coding sequence 2147–2191HSV•Tag ® coding sequence 2378–2413His•Tag ® coding sequence 2420–2443CITE sequence 2505–3004Rabbit globin terminator region 3861–4103T7 terminator 4104–4151pUC origin 5080bla coding sequence 5679–6539PshA I Blunt Cloning SitePshA IGAT GAC GAC GAC AAG AGT CCGD D D D K S PEnterokinasePshA I vectorvectorinsertGAT GAC GAC GAC AA A/G XXXinsertD D D D K codonEnterokinaseSma I Blunt Cloning SiteSma ICTG GTC CCC CGG GGC AGCL V P R G SThrombinSma I vectorvectorinsertCTG GTC CCC C GX XXXinsertL V P R codonThrombinSgf I (4874)Swa I (4418)Bpm I (5822)1629 (4165-4870)Eco47 III (4286)Sph I (4162)ori (5080)Rca I (3928)Cla I (3892)Bcg I (6291)Sca I (6232)Ap(5679-6539)T7terminatorRabbit β globinterminator<strong>pTriEx</strong>-4 <strong>Neo</strong>(6601 bp)neoExon 2(3040-3843)Nsi I (184)Mlu I (399)Bpu10 I (416)lef-2, 603 (45-1011)CMV ieenhancer& promoterExon 1T7 promoterlac operatorp10 promoterCITEAvr II (2582)BsmB I (949)Fse I (1017)Nco I (2112)Xcm I (2138)Sma I (2216)PshA I (2256)BseR I (2260)Sac II (1746) EcoR V (2280)Xba I (1815)Psp5 II (1989)Pac I (2061)Ecl136 II (2287)BamH I (2294)EcoR I (2299)Bgl II (2307)Asc I (2315)BssH II (2315)Sse8387 I (2325)PinA I (2337)Nsp V (2342)Not I (2352)Pvu II (2363)Bst1107 I (2367)Xho I (2414)Bsu36 I (2456)TB299 02/01The <strong>pTriEx</strong>-4 <strong>Neo</strong> vector 1 (Cat. No. 70933-3) is designed to allow rapid characterization oftarget genes in E. coli, insect and vertebrate cells, and to allow rapid selection of stable transfectedvertebrate cells expressing high levels of target gene. Expression in vertebrate cells is mediated bythe CMV immediate early enhancer and promoter 2 . The drug selection marker is expressed underthe control of the EMC virus Cap-Independent Translation Enhancer (CITE) sequence (or IRES), allowingrapid selection of transfected vertebrate cells using the drug G 418 or neomycin sulfate. Forexpression in insect cells, <strong>pTriEx</strong>-4 <strong>Neo</strong> contains flanking baculovirus sequences to permit the generationof recombinant baculoviruses using the Bac<strong>Vector</strong> ® System. In baculovirus-infected insectcells, expression is driven by the very late p10 promoter. Expression in E. coli is regulated by thetightly controlled T7lac promoter. Expression can be induced in hosts such as NovaBlue by infectingwith λCE6, a phage that constitutively expresses T7 RNA polymerase from the λp L and λp I promoters.Alternatively, <strong>pTriEx</strong> recombinant plasmids can be transferred into a (DE3)pLacI host thatallows induction with IPTG. Native protein can be expressed by cloning into the Nco I site, or generatedby cloning into the PshA I or Sma I sites and cleaving the fusion protein with enterokinase orthrombin, respectively.1 patent pending2 The CMV promoter is covered by U.S. Patent nos. 5,168,062 and 5,385,839 issued to the University of IowaResearch Foundation and is licensed for research use only.Tth111 II (3295)Msc I (3259)Nar I (3177)Ehe I (3178)CMV immediate early enhancer/promoter region TATA box 5 CapCAAAATGTCGTAATAACCCCGCCCCGTTGACGCAAATGGGCGGTAGGCGTGTACGGTGGGAGGTCTATATAAGCAGACGTCGTTTAGTGAACCGTCAGATCACTAGATGCTTTATTGCGGTAGTTTATCACAGTTAAATTGCTAACGCCAintronTriExUP primer #70846-3T7 promoterGTCTCGAACTTAACGTGCAGAAGTTGGTCGTGAGGCACTGGGCAGGT...160bp...AGCTCCTGGGCAACGTGCTGGTTATTGTGCTGTCTCATCATTTTGGCAAAGAATTGGATCGGACCGAAATTAATACGACTCACTATAGGGGExon 1 Exon 2lac operatorp10 promoter region p10 mRNA initiationAATTGTGAGCGGATAACAATTCCCCGGAGTTAATCCGGGACCTTTAATTCAACCCAACACAATATATTATAGTTAAATAAGAATTATTATCAAATCATTTGTATATTAATTAAAATACTATACTGTAAATTACATTTTATTTACAATCAExon 2KozakconsensusRBS Nco I His•Tag Xcm I S•Tag 18mer primer #70828-3S•Tag Sma IAAGGAGATATACCATGGCACACCATCACCACCATCACTCTTCTGGTAAAGAAACCGCTGCTGCGAAATTTGAACGCCAGCACATGGACTCGCCACCGCCTTCTGGTCTGGTCCCCCGGGGCAGCGCAGGTTCTGGTACGATTGATGACGACMetAlaHisHisHisHisHisHisSerSerGlyLysGluThrAlaAlaAlaLysPheGluArgGlnHisMetAspSerProProProSerGlyLeuValProArgGlySerAlaGlySerGlyThrIleAspAspAspExon 2thrombinenterokinaseEcl136 IIBssH IINsp VPshA IBseR I EcoR V (Sac I) BamH I EcoR I Bgl II Asc I Sse8387 IPinA INot I Pvu II Bst1107 IHSV•TagGACAAGAGTCCGGGCTTCTCCTCAACGATATCTGAGCTCGTGGATCCGAATTCTCAGATCTCGGCGCGCCTGCAGGTCGACGGTACCGGTTCGAAGCTTGCGGCCGCACAGCTGTATACACGTGCAAGCCAGCCAGAACTCGCCCCGAspLysSerProGlyPheSerSerThrIleSerGluLeuValAspProAsnSerGlnIleSerAlaArgLeuGlnValAspGlyThrGlySerLysLeuAlaAlaAlaGlnLeuTyrThrArgAlaSerGlnProGluLeuAlaProenterokinaseExon 2HSV•Tag Xho IHis•TagBsu36 ITriExDOWN primer #70847-3GAAGACCCCGAGGATCTCGAGCACCACCATCACCATCACCATCACTAAGTGATTAACCTCAGGTGCAGGCTGCCTATCAGAAGGTGGTGGCTGGTGTGG...1363bp CITE-neo...CCAATGCCCTGGCTCACAAATACCACTGAGGluAspProGluAspLeuGluHisHisHisHisHisHisHisHisEndpolyA Exon 2TriExDOWN primer #70847-3signalATCGATCTTTTTCCCTCTGCCAAAAATTATGGGGACATCATGAAGCCCCTTGAGCATCTGACTTCTGGCTAATAAAGGAAATTTATTTTCATTGCAATAGTGTGTTGGAATTTTTTGTGTCTCTCACTCGGAAGGACATATGGGAGGGCAExon 2<strong>pTriEx</strong>-4 <strong>Neo</strong> cloning/expression regionNovagen • ORDERING 800-526-7319 • TECHNICAL SUPPORT 800-207-0144
<strong>pTriEx</strong>-4 <strong>Neo</strong> Restriction SitesTB299 02/01Enzyme # Sites LocationsAatII 6 1140 1193 1276 1462 15834609AccI 3 244 2328 2366AciI 61AflIII 7 399 2369 2746 2921 46794829 5018AhdI 2 499 5752AluI 21Alw26I 8 285 949 1449 1659 18974014 5813 6589AlwI 16AlwNI 3 1865 2231 5433ApaI 2 1712 2548ApaLI 3 2908 5331 6419ApoI 12AscI 1 2315AvaI 4 1732 2214 2405 2414AvaII 7 1921 1989 2209 3693 42375890 6112AvrII 1 2582BamHI 1 2293BanI 7 1480 2333 2724 2872 31763211 5700BanII 4 1712 2289 2548 3542BbsI 5 498 2406 2557 2671 4787BbvI 25BcgI 1 6291BfaI 12BglI 5 1105 1227 1298 2733 5872BglII 1 2307BpmI 1 5822Bpu10I 1 416BsaAI 5 1355 2372 2747 3481 4328BsaHI 10 495 1137 1190 1273 14591580 3177 4606 4806 6289BsaI 2 285 5813BsaJI 17BsaWI 7 833 2336 2502 3208 52235370 6043BseRI 1 2260BsgI 2 1689 2482BsiEI 7 2355 3086 4874 4934 53576122 6271BsiHKAI 8 2289 2421 2912 3290 34805335 6338 6423BslI 19BsmBI 1 949BsmFI 11BsmI 2 2577 2616Bsp1286I 13BspLU11I 2 2921 5018BspMI 6 1686 2216 2314 2770 30643445BsrBI 4 1728 1960 3790 4951BsrDI 6 79 2542 3410 3979 58135987BsrFI 8 42 781 1013 1806 23363496 3677 5832BsrGI 3 49 768 4658BsrI 16BssHII 1 2315BssSI 4 2288 3769 5190 6416Bst1107I 1 2367BstXI 2 167 3868BstYI 9 2293 2307 2410 2888 33485658 5669 6377 6394Bsu36I 1 2456Cac8I 29ClaI 1 3892CviJI 101DdeI 10 416 2283 2304 2443 24563840 3886 5292 5709 6249Enzyme # Sites LocationsDpnI 29DraI 4 429 4048 4418 6329DraIII 2 2446 2794DrdI 2 3204 5125DsaI 3 1743 2112 2965EaeI 9 1011 2352 2734 3047 30833257 3648 3675 6140EagI 2 2352 3083EarI 6 51 547 2143 3521 37316547Ecl136II 1 2287Eco47III 1 4286Eco57I 5 2876 3323 3755 5565 6419EcoO109I 6 1709 1989 2544 2900 29544120EcoRI 1 2299EcoRII 11EcoRV 1 2280EheI 1 3178FauI 9 1108 1134 1301 1529 17121738 1775 3024 3300Fnu4HI 45FokI 6 2873 3501 3526 5718 58996186FseI 1 1017FspI 3 659 3279 5974HaeII 3 3180 4288 5265HaeIII 25HgaI 9 146 503 966 1541 42284388 4814 5128 6278HhaI 24HincII 3 245 1531 2329HindIII 3 2345 2655 3003HinfI 15HphI 11 183 879 2117 2420 24262821 3355 5822 6238 6444KpnI 2 2337 2876MaeIII 15MboII 21MluI 1 399MnlI 35MscI 1 3259MseI 38MslI 8 950 1380 1837 3614 40576004 6163 6522MspA1I 8 655 1745 2156 2362 27015359 5604 6387MspI 31MunI 2 4317 4662MwoI 29NarI 1 3177NciI 12NcoI 1 2112NdeI 4 1249 4028 4088 4096NgoAIV 3 781 1013 3677NlaIII 20NlaIV 21NotI 1 2352NsiI 1 184NspI 4 2916 2925 4162 5022NspV 1 2342PacI 1 2061PflMI 2 2182 2884PinAI 1 2336PleI 10 150 1419 1931 2180 22652825 3600 4661 5396 5741PmlI 2 2372 2747PshAI 1 2256Psp1406I 2 5978 6351Psp5II 1 1989PstI 3 2325 3033 3230PvuI 2 4874 6122Enzyme # Sites LocationsPvuII 1 2362RcaI 1 3928RsaI 18RsrII 2 1921 3693SacI 1 2289SacII 1 1746SalI 2 243 2327SapI 2 3521 3731Sau3AI 29Sau96I 19ScaI 1 6232ScrFI 23SfaNI 16SfcI 9 1857 1943 2321 2679 30293226 5282 5473 5993SgfI 1 4874SmaI 1 2216SnaBI 2 1355 4328SphI 1 4162Sse8387I 1 2325SspI 4 425 4422 4619 6556StyI 3 2112 2582 4115SwaI 1 4418TaiI 25TaqI 18TfiI 5 446 3662 3796 4291 4993ThaI 16TseI 25Tsp45I 5 1802 3297 3603 6008 6219Tsp509I 42TspRI 14Tth111I 1 3295VspI 5 1022 1930 2057 4813 5924XbaI 1 1815XcmI 1 2138XhoI 1 2414XmnI 2 2643 6351Enzymes that do not cut <strong>pTriEx</strong>-4 <strong>Neo</strong>:AflII BclI Bpu1102I BsaBI BspEIBstEII EcoNI HpaI NheI NruIPmeI SanDI SexAI SfiI SgrAISpeI SrfI StuI SunINovagen • FAX 608-238-1388 • E-MAIL novatech@novagen.com