EPICENTRE Forum 14-2
EPICENTRE Forum 14-2
EPICENTRE Forum 14-2
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
<strong>EPICENTRE</strong>® Biotechnologies <strong>Forum</strong><br />
A Simple Enzymatic Process for Generating mRNA-Enriched<br />
Poly(A)-Tailed RNA from Total RNA Samples<br />
Merriann Carey, <strong>EPICENTRE</strong> Biotechnologies<br />
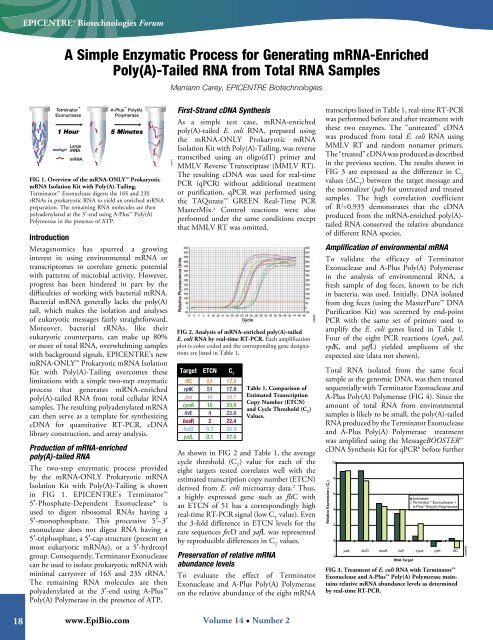
FIG 1. Overview of the mRNA-ONLY Prokaryotic<br />
mRNA Isolation Kit with Poly(A)-Tailing.<br />
Terminator Exonuclease digests the 16S and 23S<br />
rRNAs in prokaryotic RNA to yield an enriched mRNA<br />
preparation. The remaining RNA molecules are then<br />
polyadenylated at the 3′-end using A-Plus Poly(A)<br />
Polymerase in the presence of ATP.<br />
Introduction<br />
Metagenomics has spurred a growing<br />
interest in using environmental mRNA or<br />
transcriptomes to correlate genetic potential<br />
with patterns of microbial activity. However,<br />
progress has been hindered in part by the<br />
difficulties of working with bacterial mRNA.<br />
Bacterial mRNA generally lacks the poly(A)<br />
tail, which makes the isolation and analyses<br />
of eukaryotic messages fairly straightforward.<br />
Moreover, bacterial rRNAs, like their<br />
eukaryotic counterparts, can make up 80%<br />
or more of total RNA, overwhelming samples<br />
with background signals. <strong>EPICENTRE</strong>’s new<br />
mRNA-ONLY Prokaryotic mRNA Isolation<br />
Kit with Poly(A)-Tailing overcomes these<br />
limitations with a simple two-step enzymatic<br />
process that generates mRNA-enriched<br />
poly(A)-tailed RNA from total cellular RNA<br />
samples. The resulting polyadenylated mRNA<br />
can then serve as a template for synthesizing<br />
cDNA for quantitative RT-PCR, cDNA<br />
library construction, and array analysis.<br />
Production of mRNA-enriched<br />
poly(A)-tailed RNA<br />
The two-step enzymatic process provided<br />
by the mRNA-ONLY Prokaryotic mRNA<br />
Isolation Kit with Poly(A)-Tailing is shown<br />
in FIG 1. <strong>EPICENTRE</strong>’s Terminator<br />
5′-Phosphate-Dependent Exonuclease* is<br />
used to digest ribosomal RNAs having a<br />
5′-monophosphate. This processive 5′–3′<br />
exonuclease does not digest RNA having a<br />
5′-triphosphate, a 5′-cap structure (present on<br />
most eukaryotic mRNAs), or a 5′-hydroxyl<br />
group. Consequently, Terminator Exonuclease<br />
can be used to isolate prokaryotic mRNA with<br />
minimal carryover of 16S and 23S rRNA. 1<br />
The remaining RNA molecules are then<br />
polyadenylated at the 3′-end using A-Plus<br />
Poly(A) Polymerase in the presence of ATP.<br />
First-Strand cDNA Synthesis<br />
As a simple test case, mRNA-enriched<br />
poly(A)-tailed E. coli RNA, prepared using<br />
the mRNA-ONLY Prokaryotic mRNA<br />
Isolation Kit with Poly(A)-Tailing, was reverse<br />
transcribed using an oligo(dT) primer and<br />
MMLV Reverse Transcriptase (MMLV RT).<br />
The resulting cDNA was used for real-time<br />
PCR (qPCR) without additional treatment<br />
or purification. qPCR was performed using<br />
the TAQurate GREEN Real-Time PCR<br />
MasterMix. a Control reactions were also<br />
performed under the same conditions except<br />
that MMLV RT was omitted.<br />
FIG 2. Analysis of mRNA-enriched poly(A)-tailed<br />
E. coli RNA by real-time RT-PCR. Each amplification<br />
plot is color coded and the corresponding gene designations<br />
are listed in Table 1.<br />
Target ETCN C T<br />
fliC 51 17.2<br />
rplK 51 17.9<br />
pal 18 18.1<br />
cyoA 16 20.6<br />
ilvE 4 22.0<br />
basR 2 22.4<br />
fecD 0.3 25.9<br />
yafL 0.1 27.0<br />
Table 1. Comparison of<br />
Estimated Transcription<br />
Copy Number (ETCN)<br />
and Cycle Threshold (C T<br />
)<br />
Values.<br />
As shown in FIG 2 and Table 1, the average<br />
cycle threshold (C T<br />
) value for each of the<br />
eight targets tested correlates well with the<br />
estimated transcription copy number (ETCN)<br />
derived from E. coli microarray data. 2 Thus,<br />
a highly expressed gene such as fliC with<br />
an ETCN of 51 has a correspondingly high<br />
real-time RT-PCR signal (low C T<br />
value). Even<br />
the 3-fold difference in ETCN levels for the<br />
rare sequences fecD and yafL was represented<br />
by reproducible differences in C T<br />
values.<br />
Preservation of relative mRNA<br />
abundance levels<br />
To evaluate the effect of Terminator<br />
Exonuclease and A-Plus Poly(A) Polymerase<br />
on the relative abundance of the eight mRNA<br />
transcripts listed in Table 1, real-time RT-PCR<br />
was performed before and after treatment with<br />
these two enzymes. The “untreated” cDNA<br />
was produced from total E. coli RNA using<br />
MMLV RT and random nonamer primers.<br />
The “treated” cDNA was produced as described<br />
in the previous section. The results shown in<br />
FIG 3 are expressed as the difference in C T<br />
values (∆C T<br />
) between the target message and<br />
the normalizer (pal) for untreated and treated<br />
samples. The high correlation coefficient<br />
of R 2 =0.935 demonstrates that the cDNA<br />
produced from the mRNA-enriched poly(A)-<br />
tailed RNA conserved the relative abundance<br />
of different RNA species.<br />
Amplification of environmental mRNA<br />
To validate the efficacy of Terminator<br />
Exonuclease and A-Plus Poly(A) Polymerase<br />
in the analysis of environmental RNA, a<br />
fresh sample of dog feces, known to be rich<br />
in bacteria, was used. Initially, DNA isolated<br />
from dog feces (using the MasterPure DNA<br />
Purification Kit) was screened by end-point<br />
PCR with the same set of primers used to<br />
amplify the E. coli genes listed in Table 1.<br />
Four of the eight PCR reactions (cyoA, pal,<br />
rplK, and yafL) yielded amplicons of the<br />
expected size (data not shown).<br />
Total RNA isolated from the same fecal<br />
sample as the genomic DNA, was then treated<br />
sequentially with Terminator Exonuclease and<br />
A-Plus Poly(A) Polymerase (FIG 4). Since the<br />
amount of total RNA from environmental<br />
samples is likely to be small, the poly(A)-tailed<br />
RNA produced by the Terminator Exonuclease<br />
and A-Plus Poly(A) Polymerase treatment<br />
was amplified using the MessageBOOSTER<br />
cDNA Synthesis Kit for qPCR b before further<br />
Relative Expression (∆C T )<br />
10<br />
8<br />
6<br />
4<br />
2<br />
0<br />
-2<br />
yafL fecD basR ilvE cyoA rplK fliC<br />
RNA Target<br />
Untreated<br />
Terminator Exonuclease +<br />
A-Plus Poly(A) Polymerase<br />
FIG 3. Treatment of E. coli RNA with Terminator<br />
Exonuclease and A-Plus Poly(A) Polymerase maintains<br />
relative mRNA abundance levels as determined<br />
by real-time RT-PCR.<br />
c1930705<br />
18<br />
www.EpiBio.com<br />
Volume <strong>14</strong> • Number 2