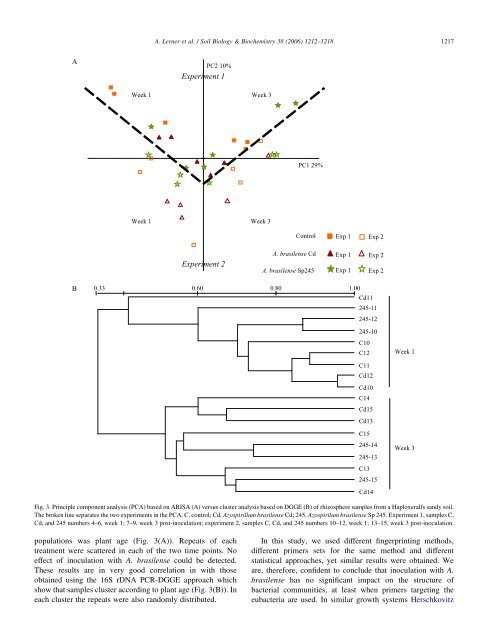
A. Lerner et al. / Soil Biology & Biochemistry 38 (2006) 1212–1218 1217 A PC2 10% Experiment 1 Week 1 Week 3 PC1 29% Week 1 Week 3 C<strong>on</strong>trol Exp 1 Exp 2 Experiment 2 A. <str<strong>on</strong>g>brasilense</str<strong>on</strong>g> Cd A. <str<strong>on</strong>g>brasilense</str<strong>on</strong>g> Sp245 Exp 1 Exp 2 Exp 1 Exp 2 B 0.33 0.60 0.80 1.00 Cd11 245-11 245-12 245-10 C10 C12 C11 Cd12 Cd10 C14 Cd15 Cd13 C15 245-14 245-13 C13 245-15 Cd14 Week 1 Week 3 Fig. 3. Principle comp<strong>on</strong>ent analysis (PCA) based <strong>on</strong> ARISA (A) versus cluster analysis based <strong>on</strong> DGGE (B) <str<strong>on</strong>g>of</str<strong>on</strong>g> rhizosphere samples from a Haploxeralfs sandy soil. The broken line separates the two experiments in the PCA. C, c<strong>on</strong>trol; Cd, <str<strong>on</strong>g>Azospirillum</str<strong>on</strong>g> <str<strong>on</strong>g>brasilense</str<strong>on</strong>g> Cd; 245, Azospirilum <str<strong>on</strong>g>brasilense</str<strong>on</strong>g> Sp 245. Experiment 1, samples C, Cd, and 245 numbers 4–6, week 1; 7–9, week 3 post-<str<strong>on</strong>g>inoculati<strong>on</strong></str<strong>on</strong>g>; experiment 2, samples C, Cd, and 245 numbers 10–12, week 1; 13–15, week 3 post-<str<strong>on</strong>g>inoculati<strong>on</strong></str<strong>on</strong>g>. populati<strong>on</strong>s was plant age (Fig. 3(A)). Repeats <str<strong>on</strong>g>of</str<strong>on</strong>g> each treatment were scattered in each <str<strong>on</strong>g>of</str<strong>on</strong>g> the two time points. No effect <str<strong>on</strong>g>of</str<strong>on</strong>g> <str<strong>on</strong>g>inoculati<strong>on</strong></str<strong>on</strong>g> with A. <str<strong>on</strong>g>brasilense</str<strong>on</strong>g> could be detected. These results are in very good correlati<strong>on</strong> in with those obtained using the 16S rDNA PCR-DGGE approach which show that samples cluster according to plant age (Fig. 3(B)). In each cluster the repeats were also randomly distributed. In this study, we used different fingerprinting methods, different primers sets for the same method and different statistical approaches, yet similar results were obtained. We are, therefore, c<strong>on</strong>fident to c<strong>on</strong>clude that <str<strong>on</strong>g>inoculati<strong>on</strong></str<strong>on</strong>g> with A. <str<strong>on</strong>g>brasilense</str<strong>on</strong>g> has no significant impact <strong>on</strong> the structure <str<strong>on</strong>g>of</str<strong>on</strong>g> bacterial communities, at least when primers targeting the eubacteria are used. In similar growth systems Herschkovitz
1218 A. Lerner et al. / Soil Biology & Biochemistry 38 (2006) 1212–1218 et al., 2005, by using the whole cell hybridizati<strong>on</strong> approach, found a high number (10 8 cfu g K1 <str<strong>on</strong>g>of</str<strong>on</strong>g> roots) <str<strong>on</strong>g>of</str<strong>on</strong>g> <str<strong>on</strong>g>Azospirillum</str<strong>on</strong>g> throughout the course <str<strong>on</strong>g>of</str<strong>on</strong>g> the experiment. Therefore, the main influence <strong>on</strong> the bacterial populati<strong>on</strong>s is due to plant development. References Baldani, V.L.D., Baldani, J.I., Dobereiner, J., 1987. Inoculati<strong>on</strong> <str<strong>on</strong>g>of</str<strong>on</strong>g> field grown wheat (Triticum aestivum) with <str<strong>on</strong>g>Azospirillum</str<strong>on</strong>g> spp. in Brasil. Biology and Fertility <str<strong>on</strong>g>of</str<strong>on</strong>g> Soils 4, 37–40. Burdman, S., Jurkevitch, E., Ok<strong>on</strong>, Y., 2000. Recent advances in the use <str<strong>on</strong>g>of</str<strong>on</strong>g> plant growth promoting rhizobacteria (PGPR) in agriculture. In: Subba Rao, N.S., Dommergues, Y.R. (Eds.), Microbial Interacti<strong>on</strong>s in Agriculture and Forestry. Science Publishers, Inc., Enfield, USA, pp. 229–250. Burdman, S., Kadouri, D., Jurkevitch, E., Ok<strong>on</strong>, Y., 2002. Bacterial phytostimulators in the rhizosphere: from research to applicati<strong>on</strong>. In: Bitt<strong>on</strong>, G. (Ed.), Encyclopedia <str<strong>on</strong>g>of</str<strong>on</strong>g> Envir<strong>on</strong>mental Microbiology, vol. 1. Wiley, L<strong>on</strong>d<strong>on</strong>, pp. 343–354. Ciccillo, F., Fiore, A., Bevivino, A., Dalmastri, C., Tabacchi<strong>on</strong>i, S., Chiarini, L., 2002. <str<strong>on</strong>g>Effect</str<strong>on</strong>g>s <str<strong>on</strong>g>of</str<strong>on</strong>g> two different applicati<strong>on</strong> methods <str<strong>on</strong>g>of</str<strong>on</strong>g> Burkholderia ambifaria MCI 7 <strong>on</strong> plant growth and rhizospheric bacterial diversity. Envir<strong>on</strong>mental Microbiology 4, 238–245. Eskew, D.L., Focht, D.D., Ting, I.P., 1977. Nitrogen fixati<strong>on</strong> denitrificati<strong>on</strong> and pleomorphic growth in highly pigmented Spirillum lip<str<strong>on</strong>g>of</str<strong>on</strong>g>erum. Canadian Journal <str<strong>on</strong>g>of</str<strong>on</strong>g> Microbiology 34, 582–585. Garcia-Martinez, J., Acinas, S.G., Ant<strong>on</strong>, A.I., Rodriguez-Valera, F., 1999. Use <str<strong>on</strong>g>of</str<strong>on</strong>g> the 16S–23S ribosomal genes spacer regi<strong>on</strong> in studies <str<strong>on</strong>g>of</str<strong>on</strong>g> prokaryotic diversity. Journal <str<strong>on</strong>g>of</str<strong>on</strong>g> Microbiological Methods 36, 55–64. German, M.A., Burdman, S., Ok<strong>on</strong>, Y., Kigel, J., 2000. <str<strong>on</strong>g>Effect</str<strong>on</strong>g>s <str<strong>on</strong>g>of</str<strong>on</strong>g> <str<strong>on</strong>g>Azospirillum</str<strong>on</strong>g> <str<strong>on</strong>g>brasilense</str<strong>on</strong>g> <strong>on</strong> root morphology <str<strong>on</strong>g>of</str<strong>on</strong>g> comm<strong>on</strong> bean (Phaseolus vulgaris L.) under different water regimes. Biology and Fertility <str<strong>on</strong>g>of</str<strong>on</strong>g> Soils 32, 259–264. Green, S.J., 2004. Shifts in Plant-associated Microbial Populati<strong>on</strong>s Mediated by Compost. PhD thesis, The Hebrew University, Jerusalem. Herschkovitz, Y., Lerner, A., Davidov, Y., Rothballer, M., Hartmann, A., Ok<strong>on</strong>, Y., Jurkevitch, E., 2005. Inoculati<strong>on</strong> with the plant growth promoting rhizobacterium <str<strong>on</strong>g>Azospirillum</str<strong>on</strong>g> <str<strong>on</strong>g>brasilense</str<strong>on</strong>g> causes little disturbance in the rhizosphere and rhizoplane <str<strong>on</strong>g>of</str<strong>on</strong>g> maize (Zea mays). Microbial Ecology. Heuer, H., Krsek, M., Baker, P., Smalla, K., Wellingt<strong>on</strong>, E.M.H., 1997. Analysis <str<strong>on</strong>g>of</str<strong>on</strong>g> actinomycete communities by specific amplificati<strong>on</strong> <str<strong>on</strong>g>of</str<strong>on</strong>g> genes encoding 16S rRNA and gel-electrophoresis separati<strong>on</strong> in denaturing gradients. Applied and Envir<strong>on</strong>mental Microbiology 63, 3233–3241. Johnsen, K., Jacobsen, C.S., Torsvik, V., 2001. Pesticide effects <strong>on</strong> bacterial diversity in agricultural soils— a review. Biology and Fertility <str<strong>on</strong>g>of</str<strong>on</strong>g> Soils 33, 443–453. Marchesi, J.R., Sato, T., Weightman, A.J., Martin, T.A., Fry, J.C., Hiom, S.J., Wade, W.G., 1998. Design and evaluati<strong>on</strong> <str<strong>on</strong>g>of</str<strong>on</strong>g> useful bacterium-specific PCR primes that amplify genes coding for bacterial 16S rRNA. Applied and Envir<strong>on</strong>mental Microbiology 64, 795–799. Marilley, L., Vogt, G., Blanc, M., Aragno, M., 1998. Bacterial diversity in the bulk soil and rhizosphere fracti<strong>on</strong>s <str<strong>on</strong>g>of</str<strong>on</strong>g> Lolium perenne and Trifolium repens as revealed by PCR restricti<strong>on</strong> analysis <str<strong>on</strong>g>of</str<strong>on</strong>g> 16S rDNA. Plant and Soil 198, 219–224. Marschner, P., Yang, C.H., Lieberei, R., Crowley, D.E., 2001. Soil and plant specific effects <strong>on</strong> bacterial community compositi<strong>on</strong> in the rhizosphere. Soil Biology & Biochemistry 33, 1437–1445. Nacamulli, C., Bevivino, A., Dalmastri, C., Tabacchi<strong>on</strong>i, S., Chiarini, L., 1997. Perturbati<strong>on</strong> <str<strong>on</strong>g>of</str<strong>on</strong>g> maize rhizosphere micr<str<strong>on</strong>g>of</str<strong>on</strong>g>lora following seed bacterizati<strong>on</strong> with Burkholderia cepacia MCI 7. FEMS Microbiology Ecology 23, 183–193. Normand, P., P<strong>on</strong>s<strong>on</strong>net, P., Nesme, X., Neyra, M., Sim<strong>on</strong>et, P., 1996. ITS analysis <str<strong>on</strong>g>of</str<strong>on</strong>g> prokaryotes. In: Akkermans, D.L., van Elsas, J.D., de Bruijn, F.J. (Eds.), Molecular Microbial Ecology Manual. Kluwer, Dordrecht, pp. 1–12. Ok<strong>on</strong>, Y., Albrecht, S.L., Burris, R.H., 1977. Methods for growing Spirillum lip<str<strong>on</strong>g>of</str<strong>on</strong>g>erum and for counting it in pure culture and in associati<strong>on</strong> with plants. Applied and Envir<strong>on</strong>mental Microbiology 33, 85–88. Ramos, H.J.O., R<strong>on</strong>cato-Maccari, L.D.B., Souza, E.M., Soares-Ramos, J.R.L., Hungria, M., Pedrosa, F.O., 2002. M<strong>on</strong>itoring <str<strong>on</strong>g>Azospirillum</str<strong>on</strong>g>–wheat interacti<strong>on</strong>s using gfp and gusA genes c<strong>on</strong>stitutively expressed from a new broad-host range vector. Journal <str<strong>on</strong>g>of</str<strong>on</strong>g> Biotechnology 97, 243–252. Ranjard, L., Polt, F., Lata, J.C., Mougel, C., Thioulouse, J., Nazaret, S., 2001. Characterizati<strong>on</strong> <str<strong>on</strong>g>of</str<strong>on</strong>g> bacterial and fungal soil communities by automated ribosomal intergenic spacer analysis fingerprints; biological and methodological variability. Applied and Envir<strong>on</strong>mental Microbiology 67, 4479–4487. Sambrook, J., Fritsch, E.F., Maniatis, T., 1989. Molecular Cl<strong>on</strong>ing, A Laboratory Manual, sec<strong>on</strong>d ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. Schallmach, E., Minz, D., Jurkevitch, E., 2000. Culture-independent detecti<strong>on</strong> <str<strong>on</strong>g>of</str<strong>on</strong>g> changes in root-associated bacterial populati<strong>on</strong>s <str<strong>on</strong>g>of</str<strong>on</strong>g> comm<strong>on</strong> bean (Phaseolus vulgaris L.) following nitrogen depleti<strong>on</strong>. Microbial Ecology 40, 309–316. Schmalenberger, A., Schwieger, F., Tebbe, C.C., 2001. <str<strong>on</strong>g>Effect</str<strong>on</strong>g> <str<strong>on</strong>g>of</str<strong>on</strong>g> primers hybridizing to different evoluti<strong>on</strong>arily c<strong>on</strong>served regi<strong>on</strong>s <str<strong>on</strong>g>of</str<strong>on</strong>g> the smallsubunit rRNA gene in PCR-based microbial community analyses and genetic pr<str<strong>on</strong>g>of</str<strong>on</strong>g>iling. Applied and Envir<strong>on</strong>mental Microbiology 67, 3557–3563. Semenov, A.M., van Bruggen, A.H.C., Zelenev, V.V., 1999. Moving waves <str<strong>on</strong>g>of</str<strong>on</strong>g> bacterial populati<strong>on</strong>s and total organic carb<strong>on</strong> al<strong>on</strong>g roots <str<strong>on</strong>g>of</str<strong>on</strong>g> wheat. Microbial Ecology 37, 116–128. Smalla, K., Wieland, G., Buchner, A., Zock, A., Parzy, A., Kaiser, J., Roskot, N., Heuer, H., Berg, G., 2001. Bulk and rhizosphere soil bacterial communities studied by denaturing gradient gel electrophoresis: plantdependent enrichment and seas<strong>on</strong>al shifts revealed. Applied and Envir<strong>on</strong>mental Microbiology 67, 4742–4751. Steenhoudt, O., Vanderleyden, J., 2000. <str<strong>on</strong>g>Azospirillum</str<strong>on</strong>g>, a free-living nitrogenfixing bacterium closely associated with grasses: genetic, biochemical and ecological aspects. FEMS Microbiology Reviews 24, 487–506. Teske, A., Sigalevich, P., Cohen, Y., Muyzer, G., 1996. Molecular identificati<strong>on</strong> <str<strong>on</strong>g>of</str<strong>on</strong>g> bacteria from coculture by denaturing gradient gel electrophoresis <str<strong>on</strong>g>of</str<strong>on</strong>g> 16S ribosomal DNA fragments as a tool for isolati<strong>on</strong> in pure cultures. Applied and Envir<strong>on</strong>mental Microbiology 62, 4210–4215. Thioulouse, J., Chessel, D., Dolédec, S., Olivier, J.M., 1997. Ade-4: a multivariate analysis and graphical display s<str<strong>on</strong>g>of</str<strong>on</strong>g>tware. Statistics and Computing 7, 75–83. Tsai, Y., Ols<strong>on</strong>, B.H., 1991. Rapid method for direct extracti<strong>on</strong> <str<strong>on</strong>g>of</str<strong>on</strong>g> DNA from soil and sediments. Applied and Envir<strong>on</strong>mental Microbiology 57, 1070–1074. Vande Broek, A., Lambrecht, M., Vanderleyden, J., 1998. Bacterial chemotactic motility is important for the initiati<strong>on</strong> <str<strong>on</strong>g>of</str<strong>on</strong>g> wheat root col<strong>on</strong>izati<strong>on</strong> by <str<strong>on</strong>g>Azospirillum</str<strong>on</strong>g> <str<strong>on</strong>g>brasilense</str<strong>on</strong>g>. Microbiology 144, 2599–2606. Vincent, J.M., 1970. A manual for the practical study <str<strong>on</strong>g>of</str<strong>on</strong>g> root nodule bacteria. IBP Handbook No. 15. Blackwell, Oxford 164 p. Watanabe, K., Kodama, Y., Harayama, S., 2001. Design and evaluati<strong>on</strong> <str<strong>on</strong>g>of</str<strong>on</strong>g> PCR primes to amplify bacterial 16S ribosomal DNA fragments used for community fingerprinting. Journal <str<strong>on</strong>g>of</str<strong>on</strong>g> Microbiological Methods 44, 253–262. Wieland, G., Neumann, R., Backhaus, H., 2001. Variati<strong>on</strong> <str<strong>on</strong>g>of</str<strong>on</strong>g> microbial communities in soil, rhizosphere, and rhizoplane in resp<strong>on</strong>se to crop species, soil type, and crop development. Applied and Envir<strong>on</strong>mental Microbiology 67, 5849–5854. Wintzingerode, F.V., Gobel, U.B., Stackebrandt, E., 1997. Determinati<strong>on</strong> <str<strong>on</strong>g>of</str<strong>on</strong>g> microbial diversity in envir<strong>on</strong>mental samples: pitfalls <str<strong>on</strong>g>of</str<strong>on</strong>g> PCR-based rRNA analysis. FEMS Microbiology Reviews 21, 213–229. Yang, C.H., Crowley, D.E., 2000. Rhizosphere microbial community structure in relati<strong>on</strong> to root locati<strong>on</strong> and plant ir<strong>on</strong> nutriti<strong>on</strong>al status. Applied and Envir<strong>on</strong>mental Microbiology 66, 345–351.