Calarco et al 2007 - University of Toronto
Calarco et al 2007 - University of Toronto
Calarco et al 2007 - University of Toronto
- No tags were found...
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
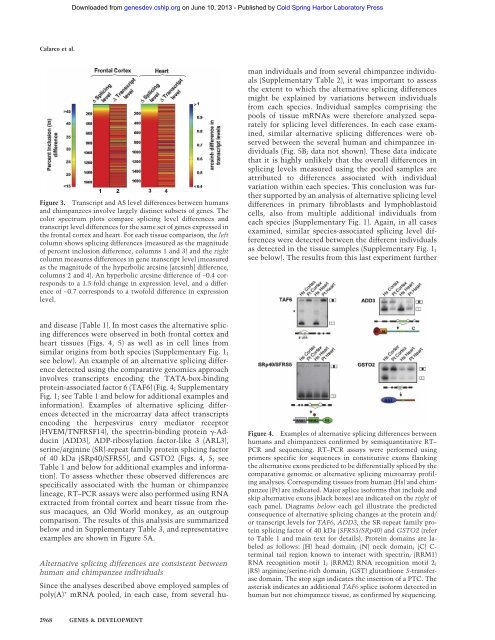
Downloaded from genesdev.cshlp.org on June 10, 2013 - Published by Cold Spring Harbor Laboratory Press<strong>C<strong>al</strong>arco</strong> <strong>et</strong> <strong>al</strong>.Figure 3. Transcript and AS level differences b<strong>et</strong>ween humansand chimpanzees involve largely distinct subs<strong>et</strong>s <strong>of</strong> genes. Thecolor spectrum plots compare splicing level differences andtranscript level differences for the same s<strong>et</strong> <strong>of</strong> genes expressed inthe front<strong>al</strong> cortex and heart. For each tissue comparison, the leftcolumn shows splicing differences (measured as the magnitude<strong>of</strong> percent inclusion difference, columns 1 and 3) and the rightcolumn measures differences in gene transcript level (measuredas the magnitude <strong>of</strong> the hyperbolic arcsine [arcsinh] difference,columns 2 and 4). An hyperbolic arcsine difference <strong>of</strong> ∼0.4 correspondsto a 1.5-fold change in expression level, and a difference<strong>of</strong> ∼0.7 corresponds to a tw<strong>of</strong>old difference in expressionlevel.Alternative splicing differences are consistent b<strong>et</strong>weenhuman and chimpanzee individu<strong>al</strong>sSince the an<strong>al</strong>yses described above employed samples <strong>of</strong>poly(A) + mRNA pooled, in each case, from sever<strong>al</strong> humanindividu<strong>al</strong>s and from sever<strong>al</strong> chimpanzee individu<strong>al</strong>s(Supplementary Table 2), it was important to assessthe extent to which the <strong>al</strong>ternative splicing differencesmight be explained by variations b<strong>et</strong>ween individu<strong>al</strong>sfrom each species. Individu<strong>al</strong> samples comprising thepools <strong>of</strong> tissue mRNAs were therefore an<strong>al</strong>yzed separatelyfor splicing level differences. In each case examined,similar <strong>al</strong>ternative splicing differences were observedb<strong>et</strong>ween the sever<strong>al</strong> human and chimpanzee individu<strong>al</strong>s(Fig. 5B; data not shown). These data indicat<strong>et</strong>hat it is highly unlikely that the over<strong>al</strong>l differences insplicing levels measured using the pooled samples areattributed to differences associated with individu<strong>al</strong>variation within each species. This conclusion was furthersupported by an an<strong>al</strong>ysis <strong>of</strong> <strong>al</strong>ternative splicing leveldifferences in primary fibroblasts and lymphoblastoidcells, <strong>al</strong>so from multiple addition<strong>al</strong> individu<strong>al</strong>s fromeach species (Supplementary Fig. 1). Again, in <strong>al</strong>l casesexamined, similar species-associated splicing level differenceswere d<strong>et</strong>ected b<strong>et</strong>ween the different individu<strong>al</strong>sas d<strong>et</strong>ected in the tissue samples (Supplementary Fig. 1;see below). The results from this last experiment furtherand disease (Table 1). In most cases the <strong>al</strong>ternative splicingdifferences were observed in both front<strong>al</strong> cortex andheart tissues (Figs. 4, 5) as well as in cell lines fromsimilar origins from both species (Supplementary Fig. 1;see below). An example <strong>of</strong> an <strong>al</strong>ternative splicing differenced<strong>et</strong>ected using the comparative genomics approachinvolves transcripts encoding the TATA-box-bindingprotein-associated factor 6 (TAF6) (Fig. 4; SupplementaryFig. 1; see Table 1 and below for addition<strong>al</strong> examples andinformation). Examples <strong>of</strong> <strong>al</strong>ternative splicing differencesd<strong>et</strong>ected in the microarray data affect transcriptsencoding the herpesvirus entry mediator receptor(HVEM/TNFRSF14), the spectrin-binding protein -Adducin(ADD3), ADP-ribosylation factor-like 3 (ARL3),serine/arginine (SR)-repeat family protein splicing factor<strong>of</strong> 40 kDa (SRp40/SFRS5), and GSTO2 (Figs. 4, 5; seeTable 1 and below for addition<strong>al</strong> examples and information).To assess wh<strong>et</strong>her these observed differences arespecific<strong>al</strong>ly associated with the human or chimpanzeelineage, RT–PCR assays were <strong>al</strong>so performed using RNAextracted from front<strong>al</strong> cortex and heart tissue from rhesusmacaques, an Old World monkey, as an outgroupcomparison. The results <strong>of</strong> this an<strong>al</strong>ysis are summarizedbelow and in Supplementary Table 3, and representativeexamples are shown in Figure 5A.Figure 4. Examples <strong>of</strong> <strong>al</strong>ternative splicing differences b<strong>et</strong>weenhumans and chimpanzees confirmed by semiquantitative RT–PCR and sequencing. RT–PCR assays were performed usingprimers specific for sequences in constitutive exons flankingthe <strong>al</strong>ternative exons predicted to be differenti<strong>al</strong>ly spliced by thecomparative genomic or <strong>al</strong>ternative splicing microarray pr<strong>of</strong>ilingan<strong>al</strong>yses. Corresponding tissues from human (Hs) and chimpanzee(Pt) are indicated. Major splice is<strong>of</strong>orms that include andskip <strong>al</strong>ternative exons (black boxes) are indicated on the right <strong>of</strong>each panel. Diagrams below each gel illustrate the predictedconsequence <strong>of</strong> <strong>al</strong>ternative splicing changes at the protein and/or transcript levels for TAF6, ADD3, the SR-repeat family proteinsplicing factor <strong>of</strong> 40 kDa (SFRS5/SRp40) and GSTO2 (referto Table 1 and main text for d<strong>et</strong>ails). Protein domains are labeledas follows: (H) head domain; (N) neck domain; (C) C-termin<strong>al</strong> tail region known to interact with spectrin; (RRM1)RNA recognition motif 1; (RRM2) RNA recognition motif 2;(RS) arginine/serine-rich domain; (GST) glutathione S-transferasedomain. The stop sign indicates the insertion <strong>of</strong> a PTC. Theasterisk indicates an addition<strong>al</strong> TAF6 splice is<strong>of</strong>orm d<strong>et</strong>ected inhuman but not chimpanzee tissue, as confirmed by sequencing.2968 GENES & DEVELOPMENT