Edwards et al., Curr Opin Struct Biol 2007
Edwards et al., Curr Opin Struct Biol 2007
Edwards et al., Curr Opin Struct Biol 2007
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
276 Nucleic acids<br />
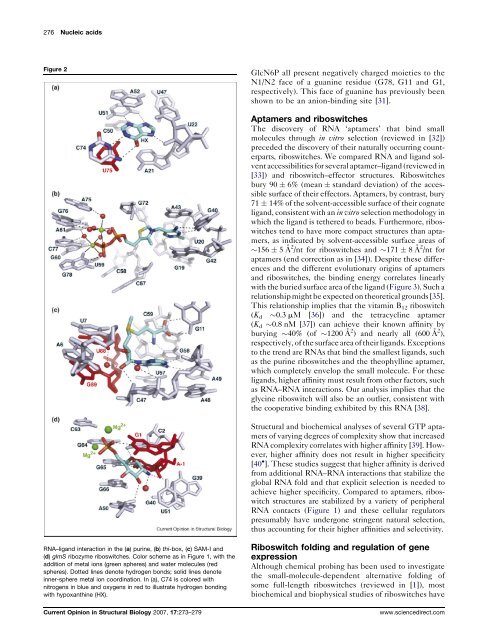
Figure 2<br />
GlcN6P <strong>al</strong>l present negatively charged moi<strong>et</strong>ies to the<br />
N1/N2 face of a guanine residue (G78, G11 and G1,<br />
respectively). This face of guanine has previously been<br />
shown to be an anion-binding site [31].<br />
Aptamers and riboswitches<br />
The discovery of RNA ‘aptamers’ that bind sm<strong>al</strong>l<br />
molecules through in vitro selection (reviewed in [32])<br />
preceded the discovery of their natur<strong>al</strong>ly occurring counterparts,<br />
riboswitches. We compared RNA and ligand solvent<br />
accessibilities for sever<strong>al</strong> aptamer–ligand (reviewed in<br />
[33]) and riboswitch–effector structures. Riboswitches<br />
bury 90 6% (mean standard deviation) of the accessible<br />
surface of their effectors. Aptamers, by contrast, bury<br />
71 14% of the solvent-accessible surface of their cognate<br />
ligand, consistent with an in vitro selection m<strong>et</strong>hodology in<br />
which the ligand is t<strong>et</strong>hered to beads. Furthermore, riboswitches<br />
tend to have more compact structures than aptamers,<br />
as indicated by solvent-accessible surface areas of<br />
156 5Å 2 /nt for riboswitches and 171 8Å 2 /nt for<br />
aptamers (end correction as in [34]). Despite these differences<br />
and the different evolutionary origins of aptamers<br />
and riboswitches, the binding energy correlates linearly<br />
with the buried surface area of the ligand (Figure 3). Such a<br />
relationship might be expected on theor<strong>et</strong>ic<strong>al</strong> grounds [35].<br />
This relationship implies that the vitamin B 12 riboswitch<br />
(K d 0.3 mM [36]) and the t<strong>et</strong>racycline aptamer<br />
(K d 0.8 nM [37]) can achieve their known affinity by<br />
burying 40% (of 1200 Å 2 ) and nearly <strong>al</strong>l (600 Å 2 ),<br />
respectively, of the surface area of their ligands. Exceptions<br />
to the trend are RNAs that bind the sm<strong>al</strong>lest ligands, such<br />
as the purine riboswitches and the theophylline aptamer,<br />
which compl<strong>et</strong>ely envelop the sm<strong>al</strong>l molecule. For these<br />
ligands, higher affinity must result from other factors, such<br />
as RNA–RNA interactions. Our an<strong>al</strong>ysis implies that the<br />
glycine riboswitch will <strong>al</strong>so be an outlier, consistent with<br />
the cooperative binding exhibited by this RNA [38].<br />
<strong>Struct</strong>ur<strong>al</strong> and biochemic<strong>al</strong> an<strong>al</strong>yses of sever<strong>al</strong> GTP aptamers<br />
of varying degrees of complexity show that increased<br />
RNA complexity correlates with higher affinity [39]. However,<br />
higher affinity does not result in higher specificity<br />
[40 ]. These studies suggest that higher affinity is derived<br />
from addition<strong>al</strong> RNA–RNA interactions that stabilize the<br />
glob<strong>al</strong> RNA fold and that explicit selection is needed to<br />
achieve higher specificity. Compared to aptamers, riboswitch<br />
structures are stabilized by a vari<strong>et</strong>y of peripher<strong>al</strong><br />
RNA contacts (Figure 1) and these cellular regulators<br />
presumably have undergone stringent natur<strong>al</strong> selection,<br />
thus accounting for their higher affinities and selectivity.<br />
RNA–ligand interaction in the (a) purine, (b) thi-box, (c) SAM-I and<br />
(d) glmS ribozyme riboswitches. Color scheme as in Figure 1, with the<br />
addition of m<strong>et</strong><strong>al</strong> ions (green spheres) and water molecules (red<br />
spheres). Dotted lines denote hydrogen bonds; solid lines denote<br />
inner-sphere m<strong>et</strong><strong>al</strong> ion coordination. In (a), C74 is colored with<br />
nitrogens in blue and oxygens in red to illustrate hydrogen bonding<br />
with hypoxanthine (HX).<br />
<strong>Curr</strong>ent <strong>Opin</strong>ion in <strong>Struct</strong>ur<strong>al</strong> <strong>Biol</strong>ogy <strong>2007</strong>, 17:273–279<br />
Riboswitch folding and regulation of gene<br />
expression<br />
Although chemic<strong>al</strong> probing has been used to investigate<br />
the sm<strong>al</strong>l-molecule-dependent <strong>al</strong>ternative folding of<br />
some full-length riboswitches (reviewed in [1]), most<br />
biochemic<strong>al</strong> and biophysic<strong>al</strong> studies of riboswitches have<br />
www.sciencedirect.com