PhD Thesis of Ina Bock - digital version - Jacobs University
PhD Thesis of Ina Bock - digital version - Jacobs University
PhD Thesis of Ina Bock - digital version - Jacobs University
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
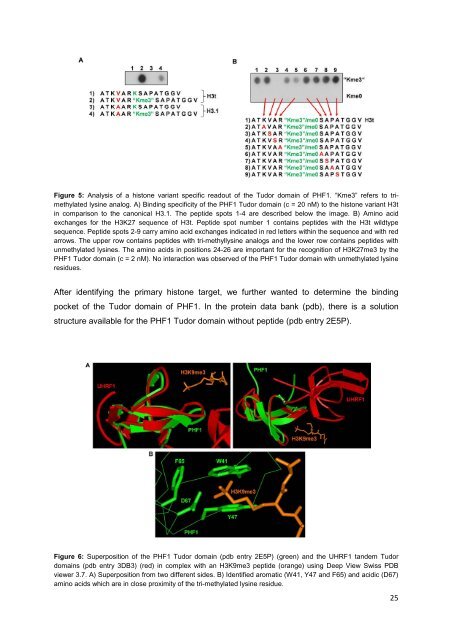
Figure 5: Analysis <strong>of</strong> a histone variant specific readout <strong>of</strong> the Tudor domain <strong>of</strong> PHF1. “Kme3” refers to trimethylated<br />
lysine analog. A) Binding specificity <strong>of</strong> the PHF1 Tudor domain (c = 20 nM) to the histone variant H3t<br />
in comparison to the canonical H3.1. The peptide spots 1-4 are described below the image. B) Amino acid<br />
exchanges for the H3K27 sequence <strong>of</strong> H3t. Peptide spot number 1 contains peptides with the H3t wildtype<br />
sequence. Peptide spots 2-9 carry amino acid exchanges indicated in red letters within the sequence and with red<br />
arrows. The upper row contains peptides with tri-methyllysine analogs and the lower row contains peptides with<br />
unmethylated lysines. The amino acids in positions 24-26 are important for the recognition <strong>of</strong> H3K27me3 by the<br />
PHF1 Tudor domain (c = 2 nM). No interaction was observed <strong>of</strong> the PHF1 Tudor domain with unmethylated lysine<br />
residues.<br />
After identifying the primary histone target, we further wanted to determine the binding<br />
pocket <strong>of</strong> the Tudor domain <strong>of</strong> PHF1. In the protein data bank (pdb), there is a solution<br />
structure available for the PHF1 Tudor domain without peptide (pdb entry 2E5P).<br />
Figure 6: Superposition <strong>of</strong> the PHF1 Tudor domain (pdb entry 2E5P) (green) and the UHRF1 tandem Tudor<br />
domains (pdb entry 3DB3) (red) in complex with an H3K9me3 peptide (orange) using Deep View Swiss PDB<br />
viewer 3.7. A) Superposition from two different sides. B) Identified aromatic (W41, Y47 and F65) and acidic (D67)<br />
amino acids which are in close proximity <strong>of</strong> the tri-methylated lysine residue.<br />
25