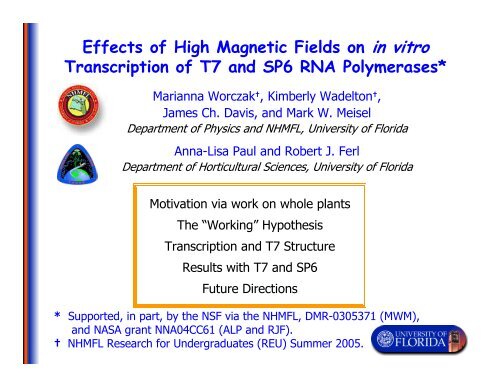
Effects of High Magnetic Fields on in vitro Transcription of T7 and ...
Effects of High Magnetic Fields on in vitro Transcription of T7 and ...
Effects of High Magnetic Fields on in vitro Transcription of T7 and ...
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
<str<strong>on</strong>g>Effects</str<strong>on</strong>g> <str<strong>on</strong>g>of</str<strong>on</strong>g> <str<strong>on</strong>g>High</str<strong>on</strong>g> <str<strong>on</strong>g>Magnetic</str<strong>on</strong>g> <str<strong>on</strong>g>Fields</str<strong>on</strong>g> <strong>on</strong> <strong>in</strong> <strong>vitro</strong><br />
Transcripti<strong>on</strong> <str<strong>on</strong>g>of</str<strong>on</strong>g> <strong>T7</strong> <strong>and</strong> SP6 RNA Polymerases*<br />
Marianna Worczak † , Kimberly Wadelt<strong>on</strong> † ,<br />
James Ch. Davis, <strong>and</strong> Mark W. Meisel<br />
Department <str<strong>on</strong>g>of</str<strong>on</strong>g> Physics <strong>and</strong> NHMFL, University <str<strong>on</strong>g>of</str<strong>on</strong>g> Florida<br />
Anna-Lisa Paul <strong>and</strong> Robert J. Ferl<br />
Department <str<strong>on</strong>g>of</str<strong>on</strong>g> Horticultural Sciences, University <str<strong>on</strong>g>of</str<strong>on</strong>g> Florida<br />
Motivati<strong>on</strong> via work <strong>on</strong> whole plants<br />
The “Work<strong>in</strong>g” Hypothesis<br />
Transcripti<strong>on</strong> <strong>and</strong> <strong>T7</strong> Structure<br />
Results with <strong>T7</strong> <strong>and</strong> SP6<br />
Future Directi<strong>on</strong>s<br />
* Supported, <strong>in</strong> part, by the NSF via the NHMFL, DMR-0305371 (MWM),<br />
<strong>and</strong> NASA grant NNA04CC61 (ALP <strong>and</strong> RJF).<br />
† NHMFL Research for Undergraduates (REU) Summer 2005.
TAGES - Plant Biom<strong>on</strong>itors <str<strong>on</strong>g>of</str<strong>on</strong>g> Microgravity<br />
An Etruscan deity who<br />
possesses wisdom. He<br />
appeared from a groove<br />
when a field was newly<br />
ploughed <strong>and</strong> taught the<br />
gathered Etruscans the<br />
skills <str<strong>on</strong>g>of</str<strong>on</strong>g> div<strong>in</strong>ati<strong>on</strong> <strong>and</strong><br />
augury.<br />
http://www.christusrex.org/www1/vaticano/ET2-Etrusco.html<br />
Transgenic<br />
Arabidopsis<br />
Gene<br />
Expressi<strong>on</strong><br />
System<br />
• Why Arabidopsis?<br />
"Arabidopsis thaliana: A Model Plant for<br />
Genome Analysis", D.W. Me<strong>in</strong>ke et al.,<br />
Science 282 (1998) 662-682.<br />
• Why Adh?<br />
Questi<strong>on</strong>s for the STS-93 experiment:<br />
Is there sufficient hypoxia <strong>in</strong> Flight to<br />
<strong>in</strong>itiate a hypoxic resp<strong>on</strong>se?<br />
Is it really hypoxia that the plants see?<br />
Adh-GUS
Adh-GUS Reporter Gene System<br />
Anoxia<br />
Hypoxia<br />
ABA<br />
Cold<br />
Drought<br />
Salt<br />
β-Glucur<strong>on</strong>idase<br />
- GUS<br />
Adh gene promoter<br />
GUS cod<strong>in</strong>g regi<strong>on</strong><br />
Quantificati<strong>on</strong> <strong>and</strong> Localizati<strong>on</strong><br />
(at the level <str<strong>on</strong>g>of</str<strong>on</strong>g> gene expressi<strong>on</strong>!)
GUS Activity<br />
[(nmol 4-MUG)/(µg prote<strong>in</strong>)/m<strong>in</strong>ute]<br />
70<br />
60<br />
20<br />
10<br />
0<br />
GUS: P = 0.001 (not B or B 2 )<br />
a Leaves (2.5 hr exposure)<br />
Run 1 - NHMFL Cell 5<br />
Run 2 - NHMFL Cell 6<br />
Run 3 - UF Superc<strong>on</strong>.<br />
Run 4 - NHMFL Cell 5<br />
Log-normal Functi<strong>on</strong><br />
95% Limits<br />
0 5 10 15 20 25<br />
<str<strong>on</strong>g>Magnetic</str<strong>on</strong>g> Field (Tesla)<br />
Real-Time Quantitative PCR<br />
Athb12 Xero2 Xtr7 Cor<br />
Microarray (8k genes)<br />
50<br />
40<br />
30<br />
20<br />
10<br />
0<br />
-10<br />
-20<br />
2.5 hrs Shoot 2.5 hrs Shoot 2.5 hrs Shoot<br />
C<strong>on</strong>trol<br />
14 T 21 T<br />
Mag C<strong>on</strong>trol Mag 14T Mag 21T<br />
The Quantitative Results (A.-L. Paul et al., MAP1 <strong>and</strong> to be published.)<br />
[Ikehata et al., J. Appl. Phys. 93 (2003) 6724: yeast <strong>in</strong> 14 T, 24 hrs: 22 genes ≤ 2 fold changes.]
The “Work<strong>in</strong>g” Hypothesis: <str<strong>on</strong>g>Magnetic</str<strong>on</strong>g> Alignment vs. Magnetophoresis<br />
The magnetic energy (Bothner-By, 1996; Tj<strong>and</strong>ra et al., 1997):<br />
E<br />
1<br />
= −<br />
2µ<br />
Variati<strong>on</strong>s from anisotropies <str<strong>on</strong>g>of</str<strong>on</strong>g> χ <strong>and</strong> B:<br />
δE<br />
= −<br />
1<br />
2µ<br />
0<br />
B(<br />
r)<br />
⋅χ⋅B(<br />
r)<br />
An order <str<strong>on</strong>g>of</str<strong>on</strong>g> magnitude comparis<strong>on</strong> <str<strong>on</strong>g>of</str<strong>on</strong>g> the two effects:<br />
Top <str<strong>on</strong>g>of</str<strong>on</strong>g> leaves <strong>and</strong> Bottom <str<strong>on</strong>g>of</str<strong>on</strong>g> roots:<br />
Anisotropy <str<strong>on</strong>g>of</str<strong>on</strong>g> χ for biomacromolecules:<br />
(Maret, Dransfeld, 1985; Tj<strong>and</strong>ra et al., 1997)<br />
(Valles et al., 1997)<br />
0<br />
{ B(<br />
r)<br />
⋅δχ<br />
⋅ B(<br />
r)<br />
+ 2 B(<br />
r)<br />
⋅ ∆χ<br />
⋅δB(<br />
r)<br />
}<br />
R<br />
≈<br />
2 ∆χ δB<br />
δχ B<br />
(δB/B) ≈ 5 × 10 −3<br />
δχ ≈ 10 −33 m 3 /molecule<br />
∆χ ≈ 10 −29 m 3 /molecule<br />
R < 10 − 2 ⇒ <str<strong>on</strong>g>Magnetic</str<strong>on</strong>g> Alignment > Magnetophoresis !
“Simplicity does not precede complexity,<br />
but follows it.” Alan J. Perlis<br />
(EPIGRAMS IN PROGRAMMING)<br />
Move from “complex” <strong>in</strong> vivo plants<br />
to a simple <strong>in</strong> <strong>vitro</strong> system!<br />
Try <strong>in</strong> <strong>vitro</strong> transcripti<strong>on</strong> <str<strong>on</strong>g>of</str<strong>on</strong>g> a s<strong>in</strong>gle process:<br />
Use <strong>T7</strong> Ribomax ® Express <strong>in</strong> <strong>vitro</strong> Transcripti<strong>on</strong> Kit ( ).<br />
Key elements:<br />
1. <strong>T7</strong> RNA polymerase is widely studied, <strong>in</strong>clud<strong>in</strong>g structure.<br />
2. Elements <str<strong>on</strong>g>of</str<strong>on</strong>g> kit are simple <strong>and</strong> pure (<strong>T7</strong> RNA polymerase,<br />
DNA template, rNTPs, <strong>and</strong> buffer).<br />
3. Protocol simple, <strong>and</strong> commercial kits available.
http://oreg<strong>on</strong>state.edu/<strong>in</strong>structi<strong>on</strong>/bb451/w<strong>in</strong>ter2005/stryer/ch28/Slide9.jpg<br />
Transcripti<strong>on</strong><br />
“F<strong>in</strong>gers”<br />
“Thumb”<br />
“Palm”<br />
“N-Term<strong>in</strong>al<br />
Doma<strong>in</strong>”<br />
D. Jeruzalmi <strong>and</strong> T.A. Steitz, EMBO J. 17 (1998) 4101 [PDB 1ARO].
Results: <strong>T7</strong> Reacti<strong>on</strong>s<br />
<strong>T7</strong> Electrophoresis Results<br />
C<strong>on</strong>trols | 4.5 Tesla | 9.0 Tesla<br />
Time (m<strong>in</strong>): 1 5 10 20 | 1 5 10 20 | 1 5 10 20<br />
2.3 kb<br />
1.1 kb<br />
I(t) / I(t = 1 m<strong>in</strong>)<br />
20<br />
15<br />
10<br />
5<br />
0<br />
0 5 10 15 20 25 30<br />
Time (m<strong>in</strong>utes)<br />
C<strong>on</strong>trol<br />
4.5 Tesla<br />
9.0 Tesla<br />
20 – 25 Tesla:<br />
“null result”?
Results: SP6 Reacti<strong>on</strong>s<br />
SP6 Electrophoresis Results<br />
C<strong>on</strong>trols | 4.5 Tesla | 9.0 Tesla<br />
1.8 kb<br />
Time (m<strong>in</strong>): 1 5 10 20 | 1 5 10 20 | 1 5 10 20<br />
Surprise aga<strong>in</strong>: 9 T str<strong>on</strong>g enough to perturb transcripti<strong>on</strong>!?<br />
SP6 RNA polymerase structure: not yet determ<strong>in</strong>ed<br />
(but believed to be similar to <strong>T7</strong> RNA polymerase).<br />
SP6: 874 residues<br />
<strong>T7</strong>: 883 residues
T 7<br />
1QLN: Cheetham <strong>and</strong> Steitz, Science 286 (1999) 2305.<br />
Model: Theis, G<strong>on</strong>g, <strong>and</strong> Mart<strong>in</strong>, Biochemistry 43 (2004) 12709.
<str<strong>on</strong>g>Effects</str<strong>on</strong>g> <str<strong>on</strong>g>of</str<strong>on</strong>g> <str<strong>on</strong>g>High</str<strong>on</strong>g> <str<strong>on</strong>g>Magnetic</str<strong>on</strong>g> <str<strong>on</strong>g>Fields</str<strong>on</strong>g> <strong>on</strong> <strong>in</strong> <strong>vitro</strong><br />
Transcripti<strong>on</strong> <str<strong>on</strong>g>of</str<strong>on</strong>g> <strong>T7</strong> <strong>and</strong> SP6 RNA Polymerases*<br />
Marianna Worczak † , Kimberly Wadelt<strong>on</strong> † ,<br />
James Ch. Davis, <strong>and</strong> Mark W. Meisel<br />
Department <str<strong>on</strong>g>of</str<strong>on</strong>g> Physics <strong>and</strong> NHMFL, University <str<strong>on</strong>g>of</str<strong>on</strong>g> Florida<br />
Anna-Lisa Paul <strong>and</strong> Robert J. Ferl<br />
Department <str<strong>on</strong>g>of</str<strong>on</strong>g> Horticultural Sciences, University <str<strong>on</strong>g>of</str<strong>on</strong>g> Florida<br />
Future Extensi<strong>on</strong>s/Work:<br />
<strong>T7</strong>/SP6: additi<strong>on</strong>al data at more/higher magnetic fields & l<strong>on</strong>ger time!<br />
(magnetic field power law?)<br />
<str<strong>on</strong>g>Magnetic</str<strong>on</strong>g> Anisotropy from Structure (Worcester, 1978; Paul<strong>in</strong>g, 1979):<br />
calculate/measure/predict differences (via RNCs) ???<br />
(RDC = residual dipolar coupl<strong>in</strong>gs)<br />
Simple to Complex: E. coli (liv<strong>in</strong>g system amplifier!) ?<br />
* Supported, <strong>in</strong> part, by the NSF via the NHMFL, DMR-0305371 (MWM),<br />
<strong>and</strong> NASA grant NNA04CC61 (ALP <strong>and</strong> RJF).<br />
† NHMFL Research for Undergraduates (REU) Summer 2005.