You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
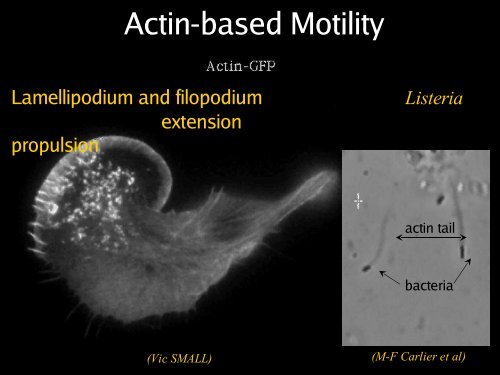
<strong>Actin</strong>-<strong>based</strong> <strong>Motility</strong><br />
Lamellipodium and filopodium<br />
extension<br />
propulsion<br />
Listeria<br />
actin tail<br />
bacteria<br />
(Vic SMALL)<br />
(M-F Carlier et al)
actin<br />
<strong>Actin</strong> Filament<br />
ATP<br />
(Ken Holmes et al, Nature 1990
Treadmilling<br />
Rate of polymerization<br />
0<br />
C cB<br />
Barbed end<br />
Pointed end<br />
concentration<br />
p<br />
T<br />
Pi<br />
e<br />
D<br />
d<br />
C ss<br />
C cP<br />
ADP<br />
ATP<br />
At steady state the filament treadmills :<br />
Subunit flux = d = e = p = k+B.(Css-CcB)
Lamellipodium Protrusion<br />
Signal<br />
TREADMILLING<br />
ADF-F-actin<br />
growing barbed end<br />
shrinking pointed end<br />
capped barbed end<br />
cross linker<br />
Anchorage<br />
nucleation<br />
elongation<br />
capping<br />
release
imicking lamellipodium with a glass rod ?<br />
30 µm Glass fibre, ∅ =1 µm<br />
<strong>Actin</strong> lamella
Treadmilling<br />
Depolymerization<br />
Polymerization<br />
Vitesse = 4µm /min = 66 nm/s = 26 a/fil/s
Synergy between Profilin and ADF<br />
ADF<br />
D<br />
D<br />
T<br />
ADF<br />
D<br />
K T prof<br />
Profilin<br />
ADP<br />
ATP<br />
Profilin<br />
K D prof<br />
Profilin<br />
T<br />
P<br />
D
Treadmilling of actin filaments :<br />
Effect of capping<br />
Rate of polymerization<br />
0<br />
C ss<br />
Barbed end<br />
C cB C cP<br />
Pointed end<br />
C ss<br />
k B + kP + (CP C -CB C )<br />
C ss<br />
C ss<br />
C ss<br />
C ss<br />
concentration<br />
Rate of filaments growth<br />
+<br />
0<br />
-<br />
J γ G<br />
0 0,5 1<br />
Extent of barbed end capping:γ<br />
« Tous pour un et un pour tous »<br />
Jγ ss<br />
J γ Les trois G mousquetaires. = 1- (1- γ γ) k B A. Dumas<br />
+ +kP +
Role of capping proteins in motility:<br />
funneling the treadmilling<br />
Slow pointed end disassembly<br />
from many capped filaments<br />
Nucleotide exchange<br />
Fast barbed end assembly<br />
of few uncapped filaments<br />
and<br />
Capping
WASP family proteins :<br />
activators of Arp2/3 complex<br />
« closed N-WASP »<br />
WH1<br />
B<br />
A<br />
GBD<br />
VV C<br />
Pro<br />
+ X<br />
+ Arp2/3<br />
+G-actin<br />
Cdc42<br />
IcsA<br />
TccP<br />
Plasma membrane<br />
« activated N-WASP »<br />
X<br />
WH1 B<br />
GBD Pro VVC A<br />
A<br />
VV C<br />
Pro<br />
R. Robinson et al<br />
(Science, 2001)<br />
(Miki et al., 1998; Machesky and Insall, 1998; Rohatgi et<br />
al., 1999; Egile et al., 1999;<br />
Kim, Rosen et al., 2000; Prehoda et al;, 2000)
Filament branching array in lamellipodia<br />
Arp 2/3<br />
50<br />
40<br />
Count<br />
30<br />
20<br />
Branching<br />
(T. Svitkina and G.G. Borisy, 1999; Blanchoin et al.; Pantaloni et al., 2000)<br />
10<br />
0<br />
0 1 2 3 4<br />
Ratio D/M
Modeling barbed end branching<br />
by Arp2/3 complex: un autocatalytic<br />
process<br />
2.5<br />
Polymerised actin, µM<br />
2<br />
1.5<br />
1<br />
0.5<br />
A+A F<br />
F +A F<br />
k +1<br />
= 2.10 -9 s -1 µM -1<br />
k -1<br />
= 5.10 -4 s -1<br />
k +2<br />
= 10 s -1 µM -1<br />
k -2<br />
= 1 s -1<br />
F +Y+2A ===> 2F<br />
Arp2/3<br />
app k +3<br />
= 0.64 s -1 µM -3<br />
0-71 nM<br />
2Y Y 2<br />
active inactive<br />
0<br />
0 500 1000 1500 2000<br />
Time, s<br />
K<br />
4<br />
= 10 -2 µM
Arp2/3: Branching Mechanism<br />
(Pantaloni et al., NCB 2000)<br />
WA or ActA
Cycles of filament attachment-detachment are coupled to branching<br />
VCA<br />
Pantaloni et al., NCB 2000; Science 2001
<strong>Actin</strong> polymerization and force production:<br />
Evolution of the Brownian Ratchet .<br />
(Oster and Mogilner, 1996-2003)
Reconstitution of actin-<strong>based</strong><br />
movement from pure proteins (Loisel et al.,<br />
Nature 1999)<br />
• Treadmilling of filaments feeds movement<br />
• Functions required for movement:<br />
3) Site-directed generation of barbed ends<br />
by N-WASP (resp. ActA)-activated Arp2/3<br />
2) Chemostat maintaining a high steady-state<br />
concentration of ATP-G-actin : <strong>Actin</strong>,<br />
ADF/cofilin, profilin, Capping protein<br />
• Movement results from a balance between the<br />
creation of new growing filaments (branching)<br />
and death of these filaments (capping).
Movement of E. coli (IcsA) and Listeria monocytogenes with<br />
pure components.<br />
E. coli IcsA<br />
Rate :<br />
≈2 µm/min<br />
Listeria<br />
Rate :<br />
≈3 µm/min<br />
Essential Proteins :<br />
N-WASP IcsA-bound<br />
Arp2/3 0.1 µM<br />
Capping Protein 0.1 µM<br />
ADF 2 µM<br />
ATP-actin+F-actin 8 µM<br />
Useful Proteins :<br />
Profilin 2 µM<br />
α-actinin 0.5 µM<br />
VASP 0.1 µM
Mimicking<br />
Lamellipodium Protrusion<br />
TREADMILLING<br />
ADF-F-actin<br />
cross linker<br />
shrinking pointed end<br />
capped barbed end
Biomimetic motility assay: actin<br />
assembly against a functionalized solid<br />
surface<br />
Beads of 3 different sizes<br />
Stuck bead
A break of symmetry in the<br />
actin meshwork leads to a<br />
polarized actin tail and<br />
movement
Biomimetic motility assay: actin assembly<br />
against a functionalized lipid membrane (GUV)
<strong>Actin</strong> tail forms and propels the liposome<br />
following break of symmetry
03:18 pm<br />
Encounters of the third kind<br />
<strong>Motility</strong> medium :<br />
N-WASP bead-bound<br />
Arp2/3 0.1 µM<br />
Capping Protein 0.1 µM<br />
ADF 2 µM<br />
ATP-actin+F-actin 8 µM<br />
Profilin 2 µM
Four Symmetric Comets
The two helices rotate in register<br />
and display opposite handedness<br />
<strong>Actin</strong> insertion
The helical parameters of the actin tails<br />
depend on the geometry of the<br />
microsphere
The surface density of N-WASP affects bead motility<br />
comet-forming beads [%]<br />
[%]<br />
100<br />
80<br />
60<br />
40<br />
20<br />
Mean velocity [µm/min]<br />
1.6<br />
1.2<br />
0.8<br />
0.4<br />
0<br />
0 5 10 15 20 25<br />
mean distance between N-WASP [nm]<br />
100<br />
• Movement requires a threshold<br />
80<br />
of N-WASP density<br />
[%]<br />
60<br />
• Velocity 40 is proportional to filament<br />
number, i.e. to N-WASP density<br />
20<br />
0<br />
0 100 200 300 400 500<br />
gelsolin concentration [nM]<br />
mean tail density [∆ gray value]<br />
0<br />
60<br />
40<br />
20<br />
0<br />
0 0.04 0.08 0.12<br />
N-WASP surface density [ molecules/nm 2 ]
Effect of external force on motility<br />
4<br />
3<br />
billes 1600 N-WASP<br />
mean velocity [ µm/min]<br />
1.6<br />
1.2<br />
0.8<br />
0.4<br />
0<br />
0 0.5 1 1.5 2 2.5 3 3.5<br />
bead diameter [µm]<br />
mean velocity [ µm/min]<br />
2<br />
1<br />
billes 200 N-WASP<br />
0<br />
0 0.2 0.4 0.6 0.8 1<br />
methyl cellulose concentration [% w/v]
Simulation of actin-<strong>based</strong> motility: balance between<br />
filament branching and capping (A.E. Carlsson, 2003,<br />
Biophys. J.)<br />
Autocatalytic branching :<br />
1. Growth velocity is independent of applied force<br />
2. Branch spacing decreases with capping
Measurement of force velocity relationship for actin-<strong>based</strong><br />
propulsion<br />
tail section versus force<br />
Experimental design<br />
force<br />
displacement<br />
pulling<br />
pushing<br />
elastic extension<br />
Tail lengthening<br />
Force velocity diagram<br />
Fast pulling, detachment and regeneration of the actin tail<br />
Marcy et al., PNAS, 2004
Arp2/3 incorporates in actin tails upon barbed<br />
end branching at the surface of N-WASP coated<br />
beads<br />
Rhodamine-actin<br />
Alexa green-Arp2/3<br />
Phase contrast<br />
Alexa green-Arp2/3<br />
Addition of Alexa green-Arp2/3
Branch spacing decreases steeply<br />
upon increasing capping (Wiesner et al.,<br />
JCB, 2003)<br />
Gelsolin: 25 nM 50 nM 100 nM 200 nM<br />
Rh-actin<br />
Alexa488<br />
-Arp2/3<br />
Both<br />
. . .<br />
. . .
Conclusions<br />
• The velocity of beads depends on the number<br />
of filaments pushing the bead.<br />
• Movement is controlled by a balance between<br />
filament branching and capping (Carlsson’s<br />
model).<br />
• Velocity is not sensitive to external load<br />
(viscous drag), i.e. the force due to<br />
polymerization at the bead surface is balanced<br />
by the internal brake (friction) due to attached<br />
filaments.
Importance of the detachment<br />
of filaments following formation<br />
of the branched junction:<br />
role of VASP
Effect of VASP on the motility of ActA-coated<br />
beads
Effect of VASP on the motility of ActA-coated<br />
beads
VASP decreases the density of<br />
filament branching<br />
+ VASP - VASP<br />
A<br />
<strong>Actin</strong><br />
Arp2/3<br />
Merged<br />
(Samarin, Romero et al., J. Cell Biol. 2003)<br />
<strong>Actin</strong>, picomoles<br />
Arp2/3, femtomoles<br />
60<br />
40<br />
20<br />
0<br />
Arp2/3<br />
(Arp2/3:actin)x1000<br />
2<br />
1<br />
0<br />
<strong>Actin</strong><br />
+ VASP VASP<br />
Arp2/3<br />
+ VASP VASP<br />
Arp2/3<br />
<strong>Actin</strong><br />
<strong>Actin</strong>
VASP enhances actin-<strong>based</strong> motility by accelerating<br />
filament detachment allowing growth after branching<br />
+ VASP<br />
ActA on BEADS<br />
Fluorescence, a.u.<br />
Soluble ActA<br />
+ VASP<br />
ActA on BEADS<br />
control<br />
0 2000 4000 6000 seconds
Without VASP the rate of detachment of the branched<br />
junction from ActA, is slow.<br />
k = 1.510 -3 s -1<br />
- VASP<br />
A<br />
YA<br />
Y<br />
A<br />
Y<br />
ActA<br />
A<br />
VASP<br />
Arp2/3<br />
Filament<br />
V<br />
Y<br />
Y<br />
A<br />
ActA-coated-Bead<br />
A<br />
Y<br />
A<br />
Y<br />
Y Y<br />
A<br />
Y<br />
- VASP<br />
k = 2.5 10 -4 s -1<br />
A<br />
Y<br />
A<br />
Y
<strong>Actin</strong>-<strong>based</strong> motility<br />
• Control of filament turnover<br />
• Site-directed generation of new filaments:<br />
2 mechanisms:<br />
Branching<br />
Barbed end<br />
nucleation (WASP/Arp2/3)<br />
and<br />
processive growth<br />
(formins)
The formin family<br />
FH3<br />
FH1<br />
FH2<br />
DAD<br />
Fmn1/2, cdc12p<br />
Rho<br />
cdc42<br />
GBD<br />
FH3<br />
FH1<br />
FH2<br />
DAD<br />
Bni1p, Bnr1p<br />
Rac1<br />
GBD<br />
FH1<br />
FH2<br />
DAD<br />
FHOS/FHOD1<br />
RhoA<br />
GBD<br />
FH3<br />
FH1<br />
FH2<br />
DAD<br />
Dia, hDia1/mDia1<br />
cdc42<br />
GBD<br />
FH3<br />
FH1<br />
FH2<br />
DAD<br />
hDia3/mDia2<br />
RhoD<br />
GBD<br />
FH3<br />
FH1<br />
FH2<br />
DAD<br />
hDia2/mDia3<br />
DAD<br />
RhoA<br />
GBD<br />
FH2<br />
FH3<br />
FH1<br />
Rho<br />
GTPases<br />
<strong>Actin</strong><br />
nucleation<br />
profilin<br />
Autoregulatory<br />
domain<br />
RhoA<br />
GBD<br />
FH3 FH1 FH2<br />
FH1<br />
DAD<br />
DAD<br />
FH2
Properties of formins<br />
• Nucleate actin assembly (FH2 is sufficient)<br />
• Active as FH2 or FH1-FH2 dimers<br />
• Bind to barbed ends without greatly affecting rat<br />
parameters for actin assembly and disassembly<br />
• Postulated to be processive « leaky cappers »<br />
remaining bound to growing barbed ends
Formin is a processive motor that directs barbed<br />
end assembly of actin filaments from profilinactin<br />
Romero et al., Cell (2004)
Formin remains bound to a growing barbed end for<br />
1200 to 2500 seconds before detaching (Romero et al.,<br />
Cell, 2004)<br />
QuickTime et un décompresseur Video sont requis pour visionner cette image.<br />
Solution of F-actin (0.5 µM) and profilin (4 µM)<br />
Frequency of detachment of a barbed end:<br />
kd = 7.5. 10 -4 ± 1.5.10-4 s -1
Formin increases the rate of profilin-actin<br />
association to barbed ends<br />
0 profilin + profilin<br />
FH1-FH2<br />
FH2<br />
free<br />
FH1-FH2<br />
free<br />
PA
Formin increases the rate of ATP hydrolysis<br />
in profilin-actin assembly
Reconstitution of formin-<strong>based</strong> motility<br />
FH1FH2<br />
coated bead<br />
ADF : 4µM<br />
Prof : 3µM<br />
F-actin : 7µM<br />
Bead velocity, µm/min<br />
20<br />
15<br />
10<br />
5<br />
0<br />
0 5 10 15 20 25 30<br />
[ADF] in µM<br />
25<br />
20<br />
15<br />
10<br />
5<br />
0<br />
0 5 10 15 20<br />
[profilin] in µM<br />
V = 2,5 µm/min
<strong>Actin</strong>-<strong>based</strong> motility<br />
LEBS, CNRS, Gif-sur-Yvette<br />
▼<br />
▼<br />
▼<br />
▼<br />
▼<br />
Dominique Pantaloni<br />
Marie-France Carlier<br />
Emmanuèle Helfer<br />
Dominique Didry<br />
Diep Lê<br />
Stanislav Samarin<br />
Sebastian Wiesner<br />
Christophe Le Clainche<br />
✆ Stéphane Romero<br />
✆ Vincent Delatour<br />
Collaborators<br />
(Institut Pasteur) :<br />
Coumaran Egile<br />
Philippe Sansonetti<br />
(Institut Curie):<br />
Jacques Prost<br />
Cécile Sykes<br />
(Harvard medical<br />
school) :<br />
Christine Kocks
Capping proteins regulate the speed and<br />
duration of formin-<strong>based</strong> motile processes
Microfilaments: Polarity,<br />
Flexibility.<br />
Persistance length : 12 µm<br />
7 nm
Mimicking « hopping Listeria »:<br />
From continuous to periodic actin<strong>based</strong><br />
movement
Formin drives rapid site-directed barbed end<br />
assembly of actin filaments from profilin-actin<br />
Distance<br />
Time
DPi<br />
DPi
ATP hydrolysis occurs on Arp2 only following<br />
branch formation and drives debranching<br />
(Le Clainche » et al., 2003, revised, PNAS)
Model for actin-<strong>based</strong> motility<br />
i n a c t i v e N - W A S P<br />
1. Signal-mediated activation of N-WASp, the branching<br />
enzyme<br />
2. G-actin, Arp2/3 and filaments as substrates of N-WASp<br />
3. Dissociation of the branch (product) and recycling<br />
Question: Role of ATP exchange and hydrolysis on Arp2/3<br />
in the branching and recycling of Arp2/3 complex ?
Dendritic nucleation on ActA<br />
beads .<br />
(Svitkina and Borisy)
Gallery
Localization of Arp2/3 complex at the branch<br />
junction and on the mother filament<br />
50<br />
40<br />
Count<br />
30<br />
20<br />
10<br />
0<br />
0 1 2 3 4<br />
Ratio D/M<br />
Rhodamine-<strong>Actin</strong><br />
200 ms<br />
Alexa488-Arp2/3<br />
30 seconds
Arp2/3-stimulated actin polymerization is an<br />
autocatalytic process<br />
2.5<br />
Polymerised actin, µM<br />
2<br />
1.5<br />
1<br />
Arp2/3<br />
0.5<br />
1-71 nM<br />
Ln [(F-Fo)/(Fmax-F)]<br />
1<br />
0<br />
-1<br />
-2<br />
-200 0 200<br />
Time, s<br />
0<br />
0 500 1000 1500 2000<br />
Time, s
Contraintes de Polymérisation<br />
?
Comète Hélicoïdale
Forces of the order of 1 nN are developed<br />
by actin polymerization<br />
Effect of methylcellulose on G(f)<br />
(Laser Tracking Microrheology)<br />
Force-velocity relationship
Using the motility assay to understand<br />
the mechanism of production of force<br />
and directional movement<br />
• Control of the concentration of soluble proteins<br />
in the motility medium.<br />
• Control of the surface density of filament<br />
branching enzyme (N-WASP or ActA).<br />
• Load/velocity relationship: control of the size of<br />
the bead and of the viscosity of the medium.<br />
• Frequency of filament branching during<br />
movement: two fluorophores (actin and Arp2/3)
<strong>Actin</strong>-<strong>based</strong> motility:<br />
How vectorial assembly of actin filaments<br />
can generate force and movement
Microfilaments: Polarité,<br />
Flexibilité.<br />
7 nm
PERSPECTIVES<br />
• Biomimetics: Reconstitution of lamellipodium protrusion<br />
(force applied to a membrane, functionalized liposome)<br />
• Coupling of adhesion and protrusion during cell<br />
migration: concerted actin dynamics at focal contacts<br />
and in lamellipodium.<br />
• Signaling, actin-<strong>based</strong> motility and morphogenesis:<br />
specifying different motile actin-<strong>based</strong> structures.
Arp2/3 Complex : downstream target of<br />
multiple signaling pathways leading to actin<br />
assembly<br />
Arp3<br />
21 47<br />
20 34<br />
16<br />
Arp2<br />
42<br />
Non-zero crosslink<br />
Two hybrid<br />
Zero-length crosslink<br />
40<br />
(Higgs and Pollard, 1999)<br />
R. Robinson et al<br />
(Science, 2001)<br />
• Localized in actin-<strong>based</strong> motility processes:<br />
• Listeria and Shigella propulsion (Welch, 1997;<br />
Egile et al., 1998)<br />
• Lamellipodium extension (Svitkina and Borisy,<br />
1999)<br />
• Phagocytic cups (Machesky, 2000)<br />
• Endocytic vesicles (Taunton et al., 1999)<br />
• <strong>Actin</strong> patches (Li 1997; Cooper 1999)<br />
• Cadherin-mediated adhesion (Yap, 2002)<br />
• Morphological events in Drosophila (Cooley,<br />
2002)<br />
• Must interact with an activator:<br />
• ActA on Listeria<br />
• WASP and Scar/WAVE proteins in
03:18 pm<br />
Encounters of the third kind<br />
<strong>Motility</strong> medium :<br />
N-WASP bead-bound<br />
Arp2/3 0.1 µM<br />
Capping Protein 0.1 µM<br />
ADF 2 µM<br />
ATP-actin+F-actin 8 µM<br />
Profilin 2 µM
Nucléation Polymérisation : f(c)<br />
Mg ++<br />
λex<br />
λem<br />
polymérisation<br />
concentration<br />
c 3<br />
c 2<br />
c 0<br />
c 1<br />
•<br />
•<br />
c 4<br />
•<br />
•<br />
•<br />
c 5<br />
•<br />
Temps, minutes
Structure de l’<strong>Actin</strong>e<br />
Séquence des 375 acides aminés<br />
constituant l’actine humaine<br />
MDDDIAALVVDNGSGMCKAGFAGDDAP<br />
RAVFPSIVGRPRHQGVMVGMGQKD<br />
SYVGDEAQSKRGILTLKYPIEHGIVTN<br />
WDDMEKIWHHTFYNELRVAPEEHP<br />
VLLTEAPLNPKANREKMTQIMFETFNT<br />
PAMYVAIQAVLSLYASGRTTGIVM<br />
DSGDGVTHTVPIYEGYALPHAILRLDL<br />
AGRDLTDYLMKILTERGYSFTTTA<br />
EREIVRDIKEKLCYVALDFEQEMATAA<br />
SSSSLEKSYELPDGQVITIGNERF<br />
RCPEALFQPSFLGMESCGIHETTFNSI<br />
MKCDVDIRKDLYANTVLSGGTTMY<br />
PGIADRMQKEITALAPSTMKIKIIAPP<br />
ERKYSVWIGGSILASLSTFQQMWI<br />
SKQEYDESGPSIVHRKCF
<strong>Actin</strong> filaments in cell movement and<br />
morphogenesis<br />
• <strong>Actin</strong> filaments have a polar structure<br />
• They are semi-flexible polymers<br />
• Assembly dynamics is regulated in vivo<br />
• Filament assembly is a dissipative<br />
reaction (hydolysis of actin-bound ATP)
Cell motility and signaling<br />
f-Met.Leu.Phe<br />
Neutrophils<br />
Chasing Leucocyte
How ATP hydrolysis regulates motility and the<br />
stability/mechanical strength of branched actin<br />
arrays<br />
Le Clainche et al., J. Biol. Chem. Accel. Publ. 2001; PNAS 2003
TP hydrolysis on Arp2/3 drives filament debranching<br />
WA<br />
Pi<br />
ADP ATP<br />
G<br />
ATP<br />
2<br />
ADP<br />
2<br />
ATP<br />
3<br />
Pi<br />
ATP<br />
G<br />
ATP<br />
2<br />
Arp2/3<br />
Activé<br />
ATP<br />
3<br />
ATP<br />
2<br />
ATP<br />
3<br />
Arp2/3<br />
Inactif
Control of actin dynamics<br />
in cell motility<br />
• Control of the [G-actin]/[F-actin] ratio<br />
• Control of filament turnover<br />
• Spatial control of the generation of new<br />
filaments (link to signaling)
Structural basis for the switch from inhibition<br />
to promotion of actin assembly in ciboulot<br />
(Hertzog et al., Cell 2004)<br />
Pointed End<br />
Barbed End
Treadmilling (Wegner, 1976)<br />
Barbed end<br />
Pointed end<br />
Pi<br />
T<br />
D<br />
C ss<br />
Css Filament turnover<br />
ADP<br />
ATP<br />
Pure actin: 0.1 µM 3µm / 90 min<br />
Lamellipodium: 2 µM ? 3 µm / 1 min
B<br />
Ciboulot regulates axonal growth in Drosophila central brain<br />
during metamorphosis by acting like profilin<br />
(Boquet et al. Cell, 2000)<br />
Low level of Cib expression<br />
Overexpression of Cib<br />
A:wild type<br />
B: mutant
Branching Models<br />
C<br />
A<br />
Autocatalytic barbed end branching<br />
B<br />
Arp2/3 activation and side branching<br />
Nucleation at the membrane and side branching
Fluorescence Video Microscopy of F-actin<br />
2000- fold dilution<br />
in unlabeled G-actin<br />
at Cc in F buffer<br />
1 filament per field<br />
= 1 filament / 50 picoliter<br />
F-Rh-actin<br />
+G-Rh- actin<br />
40 µ M<br />
F-Rh-actin<br />
20 nM<br />
+ G-actin<br />
=Cc<br />
Isambert, Venier, Maggs and Carlier (1995)
F l e xi b il i t y o f A c t i n F il ame n t s<br />
C or r e l a t io n o f t a n ge nti al di re ct io ns<br />
s<br />
θ ( s )<br />
O<br />
< c ( s ) > = c o s [ θ ( s ) - θ ( 0 ) ]<br />
< c ( s ) > = e - | s | / 2 L p<br />
l n < c ( s ) > = -<br />
s<br />
2 L p
Reconstitution of actin-<strong>based</strong><br />
movement from pure proteins<br />
Essential Proteins<br />
N-WASP<br />
Useful Proteins<br />
VASP<br />
Rate µm/min<br />
2<br />
1<br />
Arp2/3<br />
Capping<br />
protein<br />
ADF<br />
2<br />
1<br />
α-<strong>Actin</strong>in<br />
Profilin<br />
0<br />
0<br />
0.001 0.01 0.1 1 10 100.001 0.01 0.1 1 10 100<br />
Concentration, µM<br />
Concentration, µM
Motile activities of living cells<br />
Migration<br />
Endocytosis<br />
Division<br />
Intracellular<br />
transport<br />
Directionality<br />
Energy dissipation<br />
Molecular motors<br />
Directed assembly of polymer
ADF increases the treadmilling of F-actin<br />
without ADF with ADF<br />
Rate of polymerization<br />
0<br />
C cB<br />
Barbed end<br />
C cP<br />
Pointed end<br />
concentration<br />
C cP<br />
C ss<br />
Pointed end<br />
C ss
Arp2/3 interacts with barbed ends,<br />
independently of filament length<br />
(D.Pantaloni et al. 2000)<br />
Polymerised actin, µM<br />
2<br />
1<br />
0<br />
20 pM<br />
0 200 400 600<br />
Time, s<br />
control<br />
Seeds length<br />
( µm)<br />
0<br />
1<br />
2 µm<br />
Spectrin-actin<br />
Gelsolin
VASP increases the rate of detachment of the branched<br />
junction from ActA, allowing growth after branching<br />
ActA in solution<br />
V<br />
Y<br />
Y<br />
V<br />
V Y<br />
k = 1.510 -3 s -1<br />
± VASP<br />
- VASP<br />
Y<br />
V<br />
V<br />
Y<br />
Bead<br />
ActA<br />
VASP<br />
V<br />
+ VASP<br />
Arp2/3<br />
Y<br />
ActA-coated beads<br />
V<br />
Y<br />
Y V<br />
k = 3.10 -2 s -1<br />
- VASP<br />
k = 2.5 10 -4 s -1<br />
V<br />
Y<br />
+Y<br />
Filament