Wavelet basis functions in biomedical signal processing
Wavelet basis functions in biomedical signal processing
Wavelet basis functions in biomedical signal processing
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
10 J. Rafiee et al. / Expert Systems with Applications xxx (2010) xxx–xxx<br />
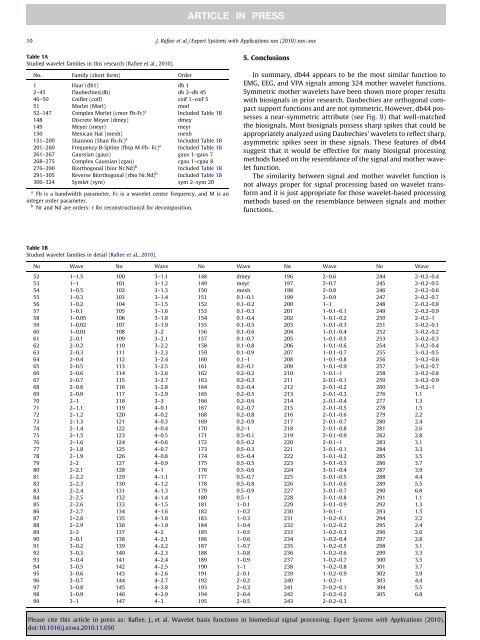
Table 1A<br />
Studied wavelet families <strong>in</strong> this research (Rafiee et al., 2010).<br />
No. Family (short form) Order<br />
1 Haar (db1) db 1<br />
2–45 Daubechies(db) db 2–db 45<br />
46–50 Coiflet (coif) coif 1–coif 5<br />
51 Morlet (Morl) morl<br />
52–147 Complex Morlet (cmor Fb-Fc) a Included Table 1B<br />
148 Discrete Meyer (dmey) dmey<br />
149 Meyer (meyr) meyr<br />
150 Mexican Hat (mexh) mexh<br />
151–200 Shannon (Shan Fb-Fc) a Included Table 1B<br />
201–260 Frequency B-Spl<strong>in</strong>e (fbsp M-Fb- Fc) a Included Table 1B<br />
261–267 Gaussian (gaus) gaus 1–gaus 7<br />
268–275 Complex Gaussian (cgau) cgau 1–cgau 8<br />
276–290 Biorthogonal (bior Nr.Nd) b Included Table 1B<br />
291–305 Reverse Biorthogonal (rbio Nr.Nd) b Included Table 1B<br />
306–324 Symlet (sym) sym 2–sym 20<br />
a<br />
Fb is a bandwidth parameter, Fc is a wavelet center frequency, and M is an<br />
<strong>in</strong>teger order parameter.<br />
b Nr and Nd are orders: r for reconstruction/d for decomposition.<br />
5. Conclusions<br />
In summary, db44 appears to be the most similar function to<br />
EMG, EEG, and VPA <strong>signal</strong>s among 324 mother wavelet <strong>functions</strong>.<br />
Symmetric mother wavelets have been shown more proper results<br />
with bio<strong>signal</strong>s <strong>in</strong> prior research. Daubechies are orthogonal compact<br />
support <strong>functions</strong> and are not symmetric. However, db44 possesses<br />
a near-symmetric attribute (see Fig. 9) that well-matched<br />
the bio<strong>signal</strong>s. Most bio<strong>signal</strong>s possess sharp spikes that could be<br />
appropriately analyzed us<strong>in</strong>g Daubechies’ wavelets to reflect sharp,<br />
asymmetric spikes seen <strong>in</strong> these <strong>signal</strong>s. These features of db44<br />
suggest that it would be effective for many bio<strong>signal</strong> process<strong>in</strong>g<br />
methods based on the resemblance of the <strong>signal</strong> and mother wavelet<br />
function.<br />
The similarity between <strong>signal</strong> and mother wavelet function is<br />
not always proper for <strong>signal</strong> process<strong>in</strong>g based on wavelet transform<br />
and it is just appropriate for those wavelet-based process<strong>in</strong>g<br />
methods based on the resemblance between <strong>signal</strong>s and mother<br />
<strong>functions</strong>.<br />
Table 1B<br />
Studied wavelet families <strong>in</strong> detail (Rafiee et al., 2010).<br />
No Wave No Wave No Wave No Wave No Wave<br />
52 1–1.5 100 3–1.1 148 dmey 196 2–0.6 244 2–0.2–0.4<br />
53 1–1 101 3–1.2 149 meyr 197 2–0.7 245 2–0.2–0.5<br />
54 1–0.5 102 3–1.3 150 mexh 198 2–0.8 246 2–0.2–0.6<br />
55 1–0.3 103 3–1.4 151 0.1–0.1 199 2–0.9 247 2–0.2–0.7<br />
56 1–0.2 104 3–1.5 152 0.1–0.2 200 1–1 248 2–0.2–0.8<br />
57 1–0.1 105 3–1.6 153 0.1–0.3 201 1–0.1–0.1 249 2–0.2–0.9<br />
58 1–0.05 106 3–1.8 154 0.1–0.4 202 1–0.1–0.2 250 2–0.2–1<br />
59 1–0.02 107 3–1.9 155 0.1–0.5 203 1–0.1–0.3 251 3–0.2–0.1<br />
60 1–0.01 108 3–2 156 0.1–0.6 204 1–0.1–0.4 252 3–0.2–0.2<br />
61 2–0.1 109 3–2.1 157 0.1–0.7 205 1–0.1–0.5 253 3–0.2–0.3<br />
62 2–0.2 110 3–2.2 158 0.1–0.8 206 1–0.1–0.6 254 3–0.2–0.4<br />
63 2–0.3 111 3–2.3 159 0.1–0.9 207 1–0.1–0.7 255 3–0.2–0.5<br />
64 2–0.4 112 3–2.4 160 0.1–1 208 1–0.1–0.8 256 3–0.2–0.6<br />
65 2–0.5 113 3–2.5 161 0.2–0.1 209 1–0.1–0.9 257 3–0.2–0.7<br />
66 2–0.6 114 3–2.6 162 0.2–0.2 210 1–0.1–1 258 3–0.2–0.8<br />
67 2–0.7 115 3–2.7 163 0.2–0.3 211 2–0.1–0.1 259 3–0.2–0.9<br />
68 2–0.8 116 3–2.8 164 0.2–0.4 212 2–0.1–0.2 260 3–0.2–1<br />
69 2–0.9 117 3–2.9 165 0.2–0.5 213 2–0.1–0.3 276 1.1<br />
70 2–1 118 3–3 166 0.2–0.6 214 2–0.1–0.4 277 1.3<br />
71 2–1.1 119 4–0.1 167 0.2–0.7 215 2–0.1–0.5 278 1.5<br />
72 2–1.2 120 4–0.2 168 0.2–0.8 216 2–0.1–0.6 279 2.2<br />
73 2–1.3 121 4–0.3 169 0.2–0.9 217 2–0.1–0.7 280 2.4<br />
74 2–1.4 122 4–0.4 170 0.2–1 218 2–0.1–0.8 281 2.6<br />
75 2–1.5 123 4–0.5 171 0.5–0.1 219 2–0.1–0.9 282 2.8<br />
76 2–1.6 124 4–0.6 172 0.5–0.2 220 2–0.1–1 283 3.1<br />
77 2–1.8 125 4–0.7 173 0.5–0.3 221 3–0.1–0.1 284 3.3<br />
78 2–1.9 126 4–0.8 174 0.5–0.4 222 3–0.1–0.2 285 3.5<br />
79 2–2 127 4–0.9 175 0.5–0.5 223 3–0.1–0.3 286 3.7<br />
80 2–2.1 128 4–1 176 0.5–0.6 224 3–0.1–0.4 287 3.9<br />
81 2–2.2 129 4–1.1 177 0.5–0.7 225 3–0.1–0.5 288 4.4<br />
82 2–2.3 130 4–1.2 178 0.5–0.8 226 3–0.1–0.6 289 5.5<br />
83 2–2.4 131 4–1.3 179 0.5–0.9 227 3–0.1–0.7 290 6.8<br />
84 2–2.5 132 4–1.4 180 0.5–1 228 3–0.1–0.8 291 1.1<br />
85 2–2.6 133 4–1.5 181 1–0.1 229 3–0.1–0.9 292 1.3<br />
86 2–2.7 134 4–1.6 182 1–0.2 230 3–0.1–1 293 1.5<br />
87 2–2.8 135 4–1.8 183 1–0.3 231 1–0.2–0.1 294 2.2<br />
88 2–2.9 136 4–1.9 184 1–0.4 232 1–0.2–0.2 295 2.4<br />
89 2–3 137 4–2 185 1–0.5 233 1–0.2–0.3 296 2.6<br />
90 3–0.1 138 4–2.1 186 1–0.6 234 1–0.2–0.4 297 2.8<br />
91 3–0.2 139 4–2.2 187 1–0.7 235 1–0.2–0.5 298 3.1<br />
92 3–0.3 140 4–2.3 188 1–0.8 236 1–0.2–0.6 299 3.3<br />
93 3–0.4 141 4–2.4 189 1–0.9 237 1–0.2–0.7 300 3.5<br />
94 3–0.5 142 4–2.5 190 1–1 238 1–0.2–0.8 301 3.7<br />
95 3–0.6 143 4–2.6 191 2–0.1 239 1–0.2–0.9 302 3.9<br />
96 3–0.7 144 4–2.7 192 2–0.2 240 1–0.2–1 303 4.4<br />
97 3–0.8 145 4–2.8 193 2–0.3 241 2–0.2–0.1 304 5.5<br />
98 3–0.9 146 4–2.9 194 2–0.4 242 2–0.2–0.2 305 6.8<br />
99 3–1 147 4–3 195 2–0.5 243 2–0.2–0.3<br />
Please cite this article <strong>in</strong> press as: Rafiee, J., et al. <strong>Wavelet</strong> <strong>basis</strong> <strong>functions</strong> <strong>in</strong> <strong>biomedical</strong> <strong>signal</strong> process<strong>in</strong>g. Expert Systems with Applications (2010),<br />
doi:10.1016/j.eswa.2010.11.050