Presentation - What is Neuro-iT.net
Presentation - What is Neuro-iT.net
Presentation - What is Neuro-iT.net
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
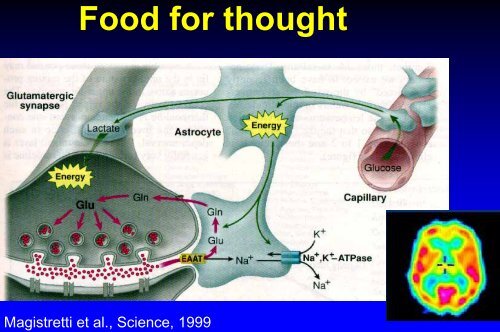
Food for thought<br />
Mag<strong>is</strong>tretti et al., Science, 1999
Hemodynamic response<br />
Blood<br />
flow<br />
<strong>Neuro</strong>electrical<br />
activity<br />
Blood flow<br />
Oxygen<br />
Hemoglobin<br />
fMRI signal<br />
Deoxyhemoglobin
Questions about the integration of<br />
EEG/MEG with the fMRI<br />
<strong>What</strong> are the techniques to usefully relate<br />
EEG/MEG and fMRI<br />
<strong>What</strong> <strong>is</strong> the evidence for true synergy<br />
<strong>What</strong> behavioral and analys<strong>is</strong> methods are<br />
successful<br />
<strong>What</strong> do we expect in the near future
Human brain produces<br />
measurable signals on the scalp<br />
Hans Berger in 1929 produced the first report<br />
on the measurement of electrical activity in<br />
man over the scalp surface<br />
He hoped that EEG could represent a sort of<br />
“window on the mind”<br />
Berger’s equipment
High Resolution<br />
Electroencephalography (HREEG)<br />
Brain activity elicited a time varying<br />
potential d<strong>is</strong>tribution over the<br />
cortical surface<br />
Such potential d<strong>is</strong>tribution are still<br />
measurable at the scalp level<br />
Due to low scalp conductivity the<br />
EEG Signal to no<strong>is</strong>e ratio <strong>is</strong> very<br />
low<br />
HREEG => Sampling the potential<br />
d<strong>is</strong>tribution with an high number of<br />
electrodes, MRI images for real<strong>is</strong>tic<br />
head modeling and spatial<br />
deblurring algorithms
teps to improve the spatial details of<br />
recorded EEG Data<br />
1975 1993<br />
1989 1996<br />
Real<strong>is</strong>tic<br />
Insert the<br />
Model Computation<br />
scalp geometry of of<br />
The of thickness skull Real<strong>is</strong>tic Head and in dura<br />
Volume Surface the mater SL in<br />
Conductor Laplacian computation<br />
inverse<br />
calculation
The neuroimaging puzzle<br />
Different neuroimaging techniques, same<br />
experimental paradigm<br />
(unilateral right middle finger movement)<br />
fMRI EEG MEG
The linear inverse problem<br />
ξ = argmin ( Ax − b +<br />
The difference between<br />
modeled and measured<br />
potentials/fields <strong>is</strong><br />
minimized, together<br />
with the energy of the<br />
sources<br />
x<br />
2<br />
M<br />
λ<br />
2<br />
A <strong>is</strong> the lead field matrix<br />
x <strong>is</strong> a vector in the source space<br />
b <strong>is</strong> the measured data vector<br />
λ <strong>is</strong> a regularization parameter<br />
M <strong>is</strong> the metric for the data space<br />
N <strong>is</strong> the metric for the source<br />
space<br />
ξ <strong>is</strong> the solution vector<br />
Solutions ξ are obtained by using x = G b where<br />
G<br />
( ) AN<br />
−1<br />
′ + λ<br />
−1<br />
1<br />
−1 −<br />
= N A′<br />
A M<br />
x<br />
2<br />
N<br />
)
Dipolar Localization Error (DLE)<br />
k-th source<br />
x<br />
Est<br />
=<br />
Gb<br />
=<br />
GAx<br />
True<br />
i-th source<br />
x<br />
Est<br />
=<br />
Rx<br />
True<br />
x = Rδ = R.<br />
i-th column of the resolution matrix R<br />
iˆ =<br />
Est<br />
DLE<br />
arg<br />
i<br />
i<br />
i<br />
index of the maximum of the<br />
max R<br />
ki<br />
k<br />
r = −<br />
r<br />
i<br />
r<br />
iˆ<br />
i-th column of the resolution matrix R<br />
d<strong>is</strong>tance between the two sources
Resolution Kernels<br />
k<br />
i<br />
j<br />
x<br />
Est<br />
i<br />
=<br />
N<br />
∑<br />
k<br />
= 1<br />
R<br />
ik<br />
x<br />
True<br />
k<br />
The R ik s define how the different sources other than the i-th<br />
contributed to the estimation to the i-th itself<br />
The R ik s belongs to the i-th row of the resolution matrix and<br />
are called Resolution Kernels
The Resolution Kernel<br />
Bad Resolution<br />
Kernel<br />
large peak around<br />
the maximum<br />
one or more peaks<br />
located far from the<br />
source position<br />
Good Resolution Kernel<br />
narrow peak around<br />
the maximum<br />
one peak located at<br />
the source position
From current strength to probability maps<br />
How obtain a measure of the uncertainty of current<br />
estimations due to the EEG/MEG no<strong>is</strong>e (n) <br />
Under the null hypothes<strong>is</strong> of<br />
no activation the Z <strong>is</strong><br />
d<strong>is</strong>tributed as a Gaussian<br />
d<strong>is</strong>tribution<br />
In the case of three<br />
component for each dipole<br />
the q as a sum of squares <strong>is</strong><br />
d<strong>is</strong>tributed as a F<strong>is</strong>her<br />
d<strong>is</strong>tribution (F 3,n )<br />
σ<br />
Z<br />
q<br />
i<br />
i<br />
2<br />
No<strong>is</strong>e<br />
( t)<br />
=<br />
( t)<br />
=<br />
=<br />
⎡<br />
⎢<br />
⎣<br />
G<br />
Gnn<br />
3<br />
k = 1<br />
3<br />
k = 1<br />
GCG<br />
∑<br />
∑<br />
G<br />
G<br />
k<br />
' G<br />
⋅ b(<br />
t)<br />
k<br />
⋅<br />
'<br />
⋅<br />
'<br />
⎤<br />
b(<br />
t)<br />
⎥<br />
⎦<br />
C<br />
=<br />
⋅<br />
GCG<br />
G<br />
k<br />
2<br />
'<br />
'
Dale et al., 2000<br />
From current strengths to<br />
probability maps<br />
Point spread functions (DLE)<br />
D<strong>is</strong>tribution of the PSF (DLE)
From current strengths to<br />
probability maps<br />
Actual dipole position<br />
Weighted minimum norm<br />
Resolution kernel<br />
No<strong>is</strong>e normalized<br />
Resolution kernel
From scalp to cortical EEG in RoIs<br />
Scalp EEG<br />
Linear inverse estimates<br />
within a RoI are collapsed<br />
(mean)<br />
M1 Hand<br />
area RoI<br />
“Virtual” electrode
Integration of EEG and MEG data<br />
EEG<br />
MEG
Integration of EEG and MEG data<br />
Why:<br />
Different sensitivities to the neural sources<br />
Increased amount of information<br />
Question:How we can fuse femtoTesla and<br />
microVolt<br />
Answer: normalizing the measures with no<strong>is</strong>e<br />
standard deviation<br />
How:<br />
Mahalanob<strong>is</strong> metric for data space<br />
Column normalization for the source<br />
space<br />
ξ = argmin( Ax−b<br />
+<br />
x<br />
2<br />
M<br />
λ<br />
2<br />
x<br />
2<br />
N<br />
)
Integration of EEG and MEG data<br />
20 ms 23 ms 18..24 ms<br />
EEG<br />
SEPs<br />
MEG<br />
SEFs<br />
EEG +<br />
MEG<br />
Fuchs et al.,<br />
EEG J., 1998
The EEG and MEG movement-related<br />
recordings
EEG, MEG and<br />
EEG/MEG indexes<br />
Liu et al., 2000<br />
Babiloni et al., 2000
Left S1 Left M1 Right S1<br />
Right M1 SMA<br />
EEG/MEG integration<br />
EEG MEG EEG/MEG<br />
EEG MEG EEG-MEG
Integration of EEG or MEG data<br />
with fMRI<br />
EEG<br />
fMRI<br />
MEG
Combining EEG and/or MEG with fMRI<br />
Why:<br />
Different spatial resolution<br />
Different time resolution<br />
• How:<br />
• Mahalanob<strong>is</strong> metric for the data space (M)<br />
• Metric on the source space (N) that takes into<br />
account:<br />
• v<strong>is</strong>ibility from the sensors (column normalization); (|| A .i || 2 )<br />
• source activity as expressed by fMRI signal α; g(α)
Dale et al., <strong>Neuro</strong>n, Vol. 26, 55–67, April, 2000,<br />
Integration of MEG and fMRI<br />
fMRI solutions<br />
MEG solutions<br />
fMRI-constrained MEG solutions
Combining EEG or MEG with fMRI<br />
Solutions ξ are obtained by using x = G b where<br />
G<br />
Proposed metric for integration of EEG, MEG and fMRI data<br />
A.<br />
i<br />
N =<br />
2<br />
=<br />
ii<br />
1+<br />
Kα<br />
Solution of the electromag<strong>net</strong>ic inverse problem with fMRI<br />
constraints when Kα >> 1<br />
Solution of electromag<strong>net</strong>ic inverse problem without fMRI<br />
constraints when Kα
Block-design fMRI signals<br />
Information on hemodynamic behaviour of<br />
cortical sources are provided on a temporal<br />
scale of minutes<br />
Diagonal metric N<br />
N<br />
4%<br />
0%<br />
Percent changes (α)<br />
− 1<br />
=<br />
ii<br />
A<br />
( α ) 2<br />
−<br />
. 2<br />
g<br />
i 2 i
Event-related fMRI signals<br />
1.03<br />
1.025<br />
1.02<br />
strength<br />
1.015<br />
1.01<br />
1.005<br />
1<br />
0.995<br />
0.99<br />
0.985<br />
EMG onset<br />
1 2 3 4 5 6 7 8<br />
seconds<br />
S1<br />
M1<br />
i j<br />
Hemodynamical behavior estimated by the correlation between the eventrelated<br />
fMRI signals from the cortical areas i and j on a seconds scale<br />
Full source metric N<br />
N<br />
ij<br />
−1<br />
=<br />
A.<br />
i<br />
2<br />
−1<br />
⋅<br />
A.<br />
j<br />
2<br />
−1<br />
⋅<br />
g<br />
( ) ( ) α ⋅ g α ⋅Corr<br />
( i,<br />
j)<br />
i<br />
j
Temporal domain:<br />
movement onset (0 msec)<br />
fMRI<br />
no fMRI diag fMRI corr fMRI<br />
4%<br />
0%<br />
Percent changes<br />
+100% Positive -100% Negative<br />
Unilateral right middle finger movement
Temporal domain:<br />
reafference peak (+110 msec)<br />
fMRI<br />
no fMRI diag fMRI corr fMRI<br />
4%<br />
0%<br />
Percent changes<br />
+100% Positive -100% Negative<br />
Unilateral right middle finger movement
Movement-related cortical dynamics<br />
HREEG<br />
HREEG<br />
+ fMRI
From current strength to probability maps<br />
How obtain a measure of the uncertainty of current<br />
estimations due to the EEG/MEG no<strong>is</strong>e (n) <br />
Under the null hypothes<strong>is</strong> of<br />
no activation the Z <strong>is</strong><br />
d<strong>is</strong>tributed as a Gaussian<br />
d<strong>is</strong>tribution<br />
In the case of three<br />
component for each dipole<br />
the q as a sum of squares <strong>is</strong><br />
d<strong>is</strong>tributed as a F<strong>is</strong>her<br />
d<strong>is</strong>tribution (F 3,n )<br />
σ<br />
Z<br />
q<br />
i<br />
i<br />
2<br />
No<strong>is</strong>e<br />
( t)<br />
=<br />
( t)<br />
=<br />
=<br />
⎡<br />
⎢<br />
⎣<br />
G<br />
Gnn<br />
3<br />
k = 1<br />
3<br />
k = 1<br />
GCG<br />
∑<br />
∑<br />
G<br />
G<br />
k<br />
' G<br />
⋅ b(<br />
t)<br />
k<br />
⋅<br />
'<br />
⋅<br />
'<br />
⎤<br />
b(<br />
t)<br />
⎥<br />
⎦<br />
C<br />
=<br />
⋅<br />
GCG<br />
G<br />
k<br />
2<br />
'<br />
'
Dale et al, <strong>Neuro</strong>n,2000<br />
Liu, 2000, PhD thes<strong>is</strong><br />
Temporal domain: from current strength<br />
to probabil<strong>is</strong>tic map<br />
n i<br />
(t)<br />
b i<br />
(t)<br />
G<br />
i-th dipole<br />
σ<br />
Z<br />
2<br />
No<strong>is</strong>ei<br />
i<br />
( t)<br />
=<br />
=<br />
[ Gnn'<br />
G'<br />
] = [ GCG'<br />
]<br />
[ G ⋅b(<br />
t)<br />
]<br />
i<br />
[ GCG'<br />
] i<br />
i<br />
i
Dale et al., 2000<br />
From current strengths to<br />
probability maps<br />
Point spread functions (DLE)<br />
D<strong>is</strong>tribution of the PSF (DLE)
From current strengths to<br />
probability maps<br />
Actual dipole position<br />
Weighted minimum norm<br />
Resolution kernel<br />
No<strong>is</strong>e normalized<br />
Resolution kernel
Movement-related cortical dynamics<br />
-0.4<br />
-5000 -4000 -3000 -2000 -1000 0 1000 2000 3000<br />
-15<br />
-5000 -4000 -3000 -2000 -1000 0 1000 2000 3000<br />
+100%<br />
Z=10<br />
t= +50 ms<br />
Current<br />
density<br />
strength<br />
0.3<br />
0.2<br />
0.1<br />
0<br />
-100%<br />
M1<br />
S1<br />
SMA<br />
S1 IPSI<br />
M1 IPSI<br />
Z=-10<br />
Z score<br />
10<br />
5<br />
0<br />
fMRIconstrained<br />
fMRIconstrained<br />
No<strong>is</strong>enormalized<br />
M1<br />
S1<br />
SMA<br />
S1 IPSI<br />
M1 IPSI<br />
-0.1<br />
-0.2<br />
-0.3<br />
-5<br />
-10
Frequency-based linear inverse source<br />
EMG onset<br />
estimation<br />
Alpha desync.<br />
average<br />
ERD<br />
b i<br />
(t)<br />
bCSD ( t)<br />
bCSD<br />
ij<br />
ii<br />
f<br />
( f ; T ) ≡ B ( f ; T ) ⋅ B ( f ; T<br />
i<br />
j<br />
*<br />
)<br />
x(<br />
t)<br />
=<br />
G ⋅b(<br />
t)<br />
G<br />
xCSD (<br />
f<br />
=<br />
⋅bCSD<br />
( f<br />
)<br />
) ⋅G′
Frequency domain: from current<br />
strength to probabil<strong>is</strong>tic map<br />
G<br />
bCSD (∆t)<br />
ii<br />
f<br />
i-th dipole<br />
σ<br />
Z<br />
2<br />
No<strong>is</strong>ei<br />
i<br />
( ∆t,<br />
= Var(<br />
bCSD<br />
f ) =<br />
ii<br />
( Τ,<br />
f ))<br />
[ G⋅bCSD<br />
( ∆t,<br />
f ) G'<br />
]<br />
σ<br />
ii<br />
No<strong>is</strong>ei<br />
i
HREEG-Movies
Conclusions<br />
High resolution EEG improved spatial details of the<br />
raw EEG potential d<strong>is</strong>tributions with respect to the<br />
standard EEG techniques<br />
Multimodal integration of high resolution EEG data<br />
with those provided by MEG and fMRI techniques <strong>is</strong><br />
possible in the framework of linear inverse problem<br />
Information about sources correlation estimated<br />
from event-related fMRI can be inserted in the<br />
solution of the linear inverse problem by using a full<br />
source metric N
Filippo Carducci<br />
Claudio Babiloni<br />
Febo Cincotti<br />
Fabio Babiloni