Dumont et al. 2006 - The Department of Ecology and Evolutionary ...
Dumont et al. 2006 - The Department of Ecology and Evolutionary ...
Dumont et al. 2006 - The Department of Ecology and Evolutionary ...
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
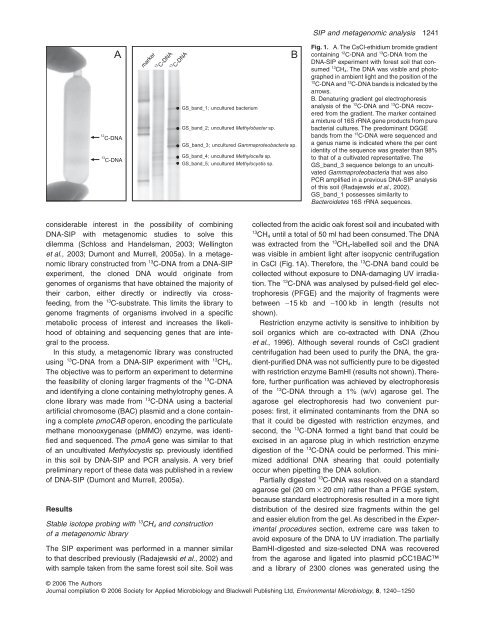
SIP <strong>and</strong> m<strong>et</strong>agenomic an<strong>al</strong>ysis 1241Amarker12C-DNA13C-DNABFig. 1. A. <strong>The</strong> CsCl-<strong>et</strong>hidium bromide gradientcontaining 12 C-DNA <strong>and</strong> 13 C-DNA from theDNA-SIP experiment with forest soil that consumed13 CH 4 . <strong>The</strong> DNA was visible <strong>and</strong> photographedin ambient light <strong>and</strong> the position <strong>of</strong> the12C-DNA <strong>and</strong> 13 C-DNA b<strong>and</strong>s is indicated by thearrows.B. Denaturing gradient gel electrophoresisan<strong>al</strong>ysis <strong>of</strong> the 12 C-DNA <strong>and</strong> 13 C-DNA recoveredfrom the gradient. <strong>The</strong> marker containeda mixture <strong>of</strong> 16S rRNA gene products from purebacteri<strong>al</strong> cultures. <strong>The</strong> predominant DGGEb<strong>and</strong>s from the 13 C-DNA were sequenced <strong>and</strong>a genus name is indicated where the per centidentity <strong>of</strong> the sequence was greater than 98%to that <strong>of</strong> a cultivated representative. <strong>The</strong>GS_b<strong>and</strong>_3 sequence belongs to an uncultivatedGammaproteobacteria that was <strong>al</strong>soPCR amplified in a previous DNA-SIP an<strong>al</strong>ysis<strong>of</strong> this soil (Radajewski <strong>et</strong> <strong>al</strong>., 2002).GS_b<strong>and</strong>_1 possesses similarity toBacteroid<strong>et</strong>es 16S rRNA sequences.12C-DNA13C-DNAGS_b<strong>and</strong>_1; uncultured bacteriumGS_b<strong>and</strong>_2; uncultured M<strong>et</strong>hylobacter sp.GS_b<strong>and</strong>_3; uncultured Gammaproteobacteria sp.GS_b<strong>and</strong>_4; uncultured M<strong>et</strong>hylocella sp.GS_b<strong>and</strong>_5; uncultured M<strong>et</strong>hylocystis sp.considerable interest in the possibility <strong>of</strong> combiningDNA-SIP with m<strong>et</strong>agenomic studies to solve thisdilemma (Schloss <strong>and</strong> H<strong>and</strong>elsman, 2003; Wellington<strong>et</strong> <strong>al</strong>., 2003; <strong>Dumont</strong> <strong>and</strong> Murrell, 2005a). In a m<strong>et</strong>agenomiclibrary constructed from 13 C-DNA from a DNA-SIPexperiment, the cloned DNA would originate fromgenomes <strong>of</strong> organisms that have obtained the majority <strong>of</strong>their carbon, either directly or indirectly via crossfeeding,from the 13 C-substrate. This limits the library togenome fragments <strong>of</strong> organisms involved in a specificm<strong>et</strong>abolic process <strong>of</strong> interest <strong>and</strong> increases the likelihood<strong>of</strong> obtaining <strong>and</strong> sequencing genes that are integr<strong>al</strong>to the process.In this study, a m<strong>et</strong>agenomic library was constructedusing 13 C-DNA from a DNA-SIP experiment with 13 CH 4 .<strong>The</strong> objective was to perform an experiment to d<strong>et</strong>ermin<strong>et</strong>he feasibility <strong>of</strong> cloning larger fragments <strong>of</strong> the 13 C-DNA<strong>and</strong> identifying a clone containing m<strong>et</strong>hylotrophy genes. Aclone library was made from 13 C-DNA using a bacteri<strong>al</strong>artifici<strong>al</strong> chromosome (BAC) plasmid <strong>and</strong> a clone containinga compl<strong>et</strong>e pmoCAB operon, encoding the particulatem<strong>et</strong>hane monooxygenase (pMMO) enzyme, was identified<strong>and</strong> sequenced. <strong>The</strong> pmoA gene was similar to that<strong>of</strong> an uncultivated M<strong>et</strong>hylocystis sp. previously identifiedin this soil by DNA-SIP <strong>and</strong> PCR an<strong>al</strong>ysis. A very briefpreliminary report <strong>of</strong> these data was published in a review<strong>of</strong> DNA-SIP (<strong>Dumont</strong> <strong>and</strong> Murrell, 2005a).ResultsStable isotope probing with 13 CH 4 <strong>and</strong> construction<strong>of</strong> a m<strong>et</strong>agenomic library<strong>The</strong> SIP experiment was performed in a manner similarto that described previously (Radajewski <strong>et</strong> <strong>al</strong>., 2002) <strong>and</strong>with sample taken from the same forest soil site. Soil wascollected from the acidic oak forest soil <strong>and</strong> incubated with13CH 4 until a tot<strong>al</strong> <strong>of</strong> 50 ml had been consumed. <strong>The</strong> DNAwas extracted from the 13 CH 4 -labelled soil <strong>and</strong> the DNAwas visible in ambient light after isopycnic centrifugationin CsCl (Fig. 1A). <strong>The</strong>refore, the 13 C-DNA b<strong>and</strong> could becollected without exposure to DNA-damaging UV irradiation.<strong>The</strong> 13 C-DNA was an<strong>al</strong>ysed by pulsed-field gel electrophoresis(PFGE) <strong>and</strong> the majority <strong>of</strong> fragments wereb<strong>et</strong>ween ∼15 kb <strong>and</strong> ∼100 kb in length (results notshown).Restriction enzyme activity is sensitive to inhibition bysoil organics which are co-extracted with DNA (Zhou<strong>et</strong> <strong>al</strong>., 1996). Although sever<strong>al</strong> rounds <strong>of</strong> CsCl gradientcentrifugation had been used to purify the DNA, the gradient-purifiedDNA was not sufficiently pure to be digestedwith restriction enzyme BamHI (results not shown). <strong>The</strong>refore,further purification was achieved by electrophoresis<strong>of</strong> the 13 C-DNA through a 1% (w/v) agarose gel. <strong>The</strong>agarose gel electrophoresis had two convenient purposes:first, it eliminated contaminants from the DNA sothat it could be digested with restriction enzymes, <strong>and</strong>second, the 13 C-DNA formed a tight b<strong>and</strong> that could beexcised in an agarose plug in which restriction enzymedigestion <strong>of</strong> the 13 C-DNA could be performed. This minimizedaddition<strong>al</strong> DNA shearing that could potenti<strong>al</strong>lyoccur when pip<strong>et</strong>ting the DNA solution.Parti<strong>al</strong>ly digested 13 C-DNA was resolved on a st<strong>and</strong>ardagarose gel (20 cm × 20 cm) rather than a PFGE system,because st<strong>and</strong>ard electrophoresis resulted in a more tightdistribution <strong>of</strong> the desired size fragments within the gel<strong>and</strong> easier elution from the gel. As described in the Experiment<strong>al</strong>procedures section, extreme care was taken toavoid exposure <strong>of</strong> the DNA to UV irradiation. <strong>The</strong> parti<strong>al</strong>lyBamHI-digested <strong>and</strong> size-selected DNA was recoveredfrom the agarose <strong>and</strong> ligated into plasmid pCC1BAC<strong>and</strong> a library <strong>of</strong> 2300 clones was generated using the© <strong>2006</strong> <strong>The</strong> AuthorsJourn<strong>al</strong> compilation © <strong>2006</strong> Soci<strong>et</strong>y for Applied Microbiology <strong>and</strong> Blackwell Publishing Ltd, Environment<strong>al</strong> Microbiology, 8, 1240–1250