Here - WM Keck Laboratory for Biological Imaging - University of ...
Here - WM Keck Laboratory for Biological Imaging - University of ...
Here - WM Keck Laboratory for Biological Imaging - University of ...
- No tags were found...
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
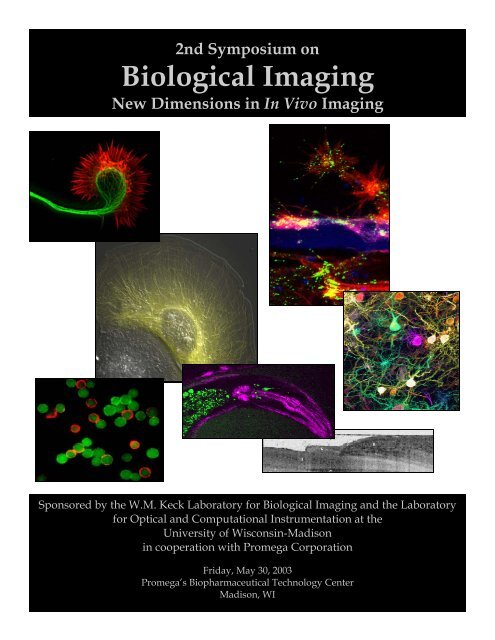
13Front cover image legend:1. Axonal growth cone <strong>of</strong> a cortical neuron treated with Netrin-1and labeled with Alexa 488 phalloidin to show actin filamentsin red and anti-tubulin antibodies to show microtubules ingreen. Netrin—1 increases filamentous actin within 30 minutes.Image courtesy <strong>of</strong> Katherine Kalil.2. A living epithelial cell expressing activated Rac1 GTPase. Thecell was injected with fluorescent tubulin and analyzed bymulti-mode DIC and fluorescence microscopy. Rac 1 activityinduces extensive microtubule growth.Image courtesy <strong>of</strong> Torsten Wittmann and Clare Waterman-Storer.3. Xenopus spinal cord explant at interface between two matrixproteins, laminin and tenascin. Red shows actin filaments labeledwith Alexa 546 phalloidin and green shows immunocytochemicallabelling <strong>of</strong> phosphotyrosine. Blue represents highestdensity <strong>of</strong> deposited laminin.Image courtesy <strong>of</strong> Timothy Gomez.4. Multicolor labeling <strong>of</strong> neurons in a mouse cortical slice usingthe "DiOlistic" technique.Image courtesy <strong>of</strong> Wen-Biao Gan.5. An OCT image <strong>of</strong> a nail fold.Image courtesy <strong>of</strong> Johannes de Boer.726546. Dual-mode non-linear optical image <strong>of</strong> the head <strong>of</strong> an adult C.elegans. A single ultrafast laser excites both second harmonicgeneration (violet) from contractile muscles and fluorescence(green) from GFP-labeled neurons and aut<strong>of</strong>luorescent granuleswithin the gut.Image courtesy <strong>of</strong> William Mohler.7. Live CHO cells were labeled with QdotTM 565 (green), 605(orange) and 655 (red) Streptavidin Conjugates. The cells weretreated with a cell-surface biotinyllation reagent be<strong>for</strong>e beingincubated with each QdotTM Streptavidin Conjugate. Then thecells labeled in different colors were mixed and examined undera epi-fluorescence microscope.Image courtesy <strong>of</strong> Xingyong Wu, Quantum Dot Corporation.
2nd Symposium on<strong>Biological</strong> <strong>Imaging</strong>New Dimensions in In Vivo <strong>Imaging</strong>Promega’s Biopharmaceutical Technology CenterMadison, WIFriday, May 30, 2003Table Of ContentsProgram Introduction Page 1Schedule <strong>of</strong> Events—Morning Session Page 2Schedule <strong>of</strong> Events—Afternoon Session Page 3Schedule <strong>of</strong> Events—Evening Activities Page 4Workshop Abstracts:Nathan O’Connor, Universal <strong>Imaging</strong> Page 5Kevin Eliceiri and LOCI Group Page 6Poster Abstracts Page 7Lecture Abstracts:Dr. William Mohler Page 11Dr. Johannes de Boer Page 12Dr. Enrico Gratton Page 13Dr. Clare Waterman-Storer Page 14Dr. Marcel Bruchez Page 15Dr. Georgyi Los Page 16Dr. Wen-Biao Gan Page 17Keynote Speaker Abstract:Dr. Mark Ellisman Page 18Vendors Page 19Sponsors Page 20Websites Page 21Notes Page 22
VWelcome to the 2nd Symposium on<strong>Biological</strong> <strong>Imaging</strong>New Dimensions in In Vivo <strong>Imaging</strong>Program Introductionisualizing the structure <strong>of</strong> living organisms has been central to biological studies from theirbeginning, with the goal <strong>of</strong> explaining the relationship between <strong>for</strong>m and function. However,living organisms are dynamic structures, and in order to understand fundamental biologicalprocesses such as the change in the con<strong>for</strong>mation <strong>of</strong> a protein, the movement and division <strong>of</strong> cells,the development <strong>of</strong> an embryo, or the integrated neural activity in an intact brain, the visualization<strong>of</strong> structures in three dimensions that change over time is necessary. Until the advent <strong>of</strong> modernbiological imaging technology it was not possible to observe directly the molecular and cellularinteractions that underlie essential functions within cells and tissues. Such interactions could only beinferred from fixed specimens. Now, however, with the aid <strong>of</strong> probes that label intracellularstructures and new imaging devices such as multiphoton microscopy, biologists can observechanges in <strong>for</strong>m related to function directly in living cells and tissues.The current explosion in our knowledge <strong>of</strong> dynamic events in molecular living organisms hasspurred a renaissance in the use and development <strong>of</strong> new imaging technology <strong>for</strong> studying live cellsand tissues. Today, probes can be made that will allow the spatial visualization <strong>of</strong> expressed geneproducts in a cell or tissue. At the single cell level, biologists can observe the dynamic movements <strong>of</strong>intracellular organelles or single vesicles at functioning neuronal synapses. Changes in thecytoskeleton <strong>of</strong> cells can be visualized directly as they occur when cells divide, migrate or executespecific functions according to their differentiated state. In tissues, the behavior <strong>of</strong> ensembles <strong>of</strong> cellsduring fundamental events in development such as gastrulation, neurulation, organogenesis, axonpathfinding and target identification, and synaptic rearrangement can be captured in real time.The symposium will highlight recent developments in in vivo imaging. Lectures and posterpresentations will demonstrate how new imaging tools can be applied to solve a variety <strong>of</strong> biologicalproblems, ranging across scale from protein structure to the intact brain. Topics covered will include:second harmonic imaging, optical coherence tomography, fluorescence correlation spectroscopy,speckle microscopy, nanocrystal labeling, small molecule reporters, and in vivo two-photonmicroscopy. In addition, interactive workshops on automated image analysis and on s<strong>of</strong>tware <strong>for</strong>biological visualization will be presented.1
<strong>Biological</strong> <strong>Imaging</strong>New Dimensions in In Vivo <strong>Imaging</strong>Schedule <strong>of</strong> EventsMorning Session7:45-8:30AM8:30AM8:45-10:45AMRegistration and Continental Breakfast-AtriumWelcome by Ron Kalil, <strong>University</strong> <strong>of</strong> Wisconsin-Madison andBob Bulleit, Promega Corporation-AuditoriumWorkshopsAutomated Methods <strong>for</strong> Light Microscopic <strong>Imaging</strong> and AnalysisNathan O’Connor, Universal <strong>Imaging</strong> CorporationRoom 236, two 1-hour sessionsFreeware and Open-Source S<strong>of</strong>tware Tools <strong>for</strong> <strong>Biological</strong> VisualizationKevin Eliceiri and LOCI Group, <strong>University</strong> <strong>of</strong> Wisconsin-MadisonRooms 216/217, two 1-hour sessions10:45-11:30AM11:30-12:30PMPoster Session and Vendor DisplaysLunch-Served buffet style in Atrium. Seating is available in Rooms104/105, 219, 221, 226/227, Cafeteria, or on the terrace (weather permitting).Seating areas are designated by posted door signage.Schedule continued on next page2
R<strong>Biological</strong> <strong>Imaging</strong>New Dimensions in In Vivo <strong>Imaging</strong>Workshop AbstractFreeware and Open-Source S<strong>of</strong>tware Tools <strong>for</strong> <strong>Biological</strong> VisualizationKevin Eliceiri and LOCI Group, <strong>University</strong> <strong>of</strong> Wisconsin-MadisonMadison, WIRooms 216/217, two 1-hour sessions8:45-9:45AM and9:45-10:45AMecent developments in non-invasive medical imaging and in vital biological microscopy haveyielded a new <strong>for</strong>m <strong>of</strong> massive data type - the multichannel, three-dimensional, time-courseimage recording. These multidimensional data sets document the dynamic changes within the fullvolume <strong>of</strong> a specimen over time, <strong>of</strong>ten simultaneously monitoring several different parameters.Examples include time-lapse, three-dimensional, light microscope image recordings and functionalMRI recordings. While the imaging modalities used to generate the data differ greatly, they share thecommon attribute <strong>of</strong> being multidimensional and are usually represented by large data archives. Tomeet the computational challenge <strong>of</strong> the effective analysis <strong>of</strong> these data, several groups, includingours, are developing s<strong>of</strong>tware visualization tools. Many <strong>of</strong> these tools are open-source applicationsthat may be readily adapted to different and emerging file <strong>for</strong>mats, and may readily adopt testedsolutions from the graphics programming community. In this workshop we will present and reviewseveral freeware and open-source tools that facilitate the incorporation <strong>of</strong> new features—demandedby new avenues <strong>of</strong> biological research—that are beyond the current scope <strong>of</strong> commercial products.6
Figure 8: Result from the different cameras in the virtual setupFigure 9: Camera positions <strong>for</strong> virtual frameworkwith a image correlation algorithm. The correlation gives us a value between the range<strong>of</strong> 0 and 1 where 0 is a bad results and 1 is the groundtruth. The table 1 presents thecorrelation values obtained <strong>for</strong> different camera position, and the figure 1 shows the 3Dmesh generated.6 CONCLUSIONS AND FUTURE WORKWe present a methodology <strong>for</strong> face reconstruction in a mixed enviroment <strong>of</strong> activepasivesetup. Structured light shows a quality improvement against the results obtainedwith pasive setups. Time multiplexing codification has the problem <strong>of</strong> motion betweenthe captured images generating a waving efect in the reconstructions. Even robustalgorithms <strong>of</strong> point matching <strong>for</strong> dense depthmaps were tested there were no real improvementin the results. We will try with color or 2D patterns which only requieres11
O<strong>Biological</strong> <strong>Imaging</strong>New Dimensions in In Vivo <strong>Imaging</strong>Lecture AbstractOptical Coherence Tomography, An Emerging <strong>Imaging</strong> Technique in MedicineJohannes de BoerAssistant Pr<strong>of</strong>essor, Harvard Medical SchoolWellman Laboratories <strong>of</strong> Photomedicine, Massachusetts General HospitalBoston, MAAuditorium1:10-1:50PMptical Coherence Tomography (OCT) is an optical imaging technique that provides crosssectional images <strong>of</strong> tissue. The technique is analogous to ultrasound, with better resolution butpoorer depth penetration. The principles <strong>of</strong> this technique will be explained, as well as functionalextensions such as Doppler OCT, which provides flow sensitivity, and Polarization Sensitive OCT(PS-OCT), which provides sensitivity to polarization properties <strong>of</strong> tissue. Advancement incomputational power allows <strong>for</strong> real time acquisition and display <strong>of</strong> Multi Functional OCT signals,and current state <strong>of</strong> the art imaging will be demonstrated. Recent results in application <strong>of</strong> thetechnique in assessing the depth <strong>of</strong> thermal injury <strong>for</strong> burn victims, screening <strong>for</strong> Basal CellCarcinoma and imaging <strong>of</strong> the retinal nerve fiber layer in ophthalmology <strong>for</strong> the early detection <strong>of</strong>Glaucoma will be discussed. A future outlook <strong>for</strong> the technology will be given.12
F<strong>Biological</strong> <strong>Imaging</strong>New Dimensions in In Vivo <strong>Imaging</strong>Lecture AbstractScanning FCS: Measuring Spatio-Temporal Correlations in the Millisecond Range inLive CellsDr. Enrico Gratton<strong>Laboratory</strong> <strong>of</strong> Fluorescence Dynamics<strong>University</strong> <strong>of</strong> Illinois at Urbana-ChampaignAuditorium1:50-2:30PMluctuation correlation spectroscopy has become a commonly used methodology to studymolecular processes in solutions and inside cells. Cross-correlation and multichannelcorrelation is also being developed in several laboratories. An important aspect <strong>of</strong> correlation whichhas not been widely exploited is the capability to detect events which occur either simultaneously orin a given statistical sequence in different spatial locations. Image correlation spectroscopy has inprinciple the capability to provide both spatial and temporal correlations. However, the commonlyused methods used <strong>for</strong> image acquisition are relatively slow and do not allow the measurement <strong>of</strong>fast correlations, typically in the millisecond scale which are typical <strong>of</strong> protein diffusion in thecytoplasm <strong>of</strong> live cells. In this work we describe a variant <strong>of</strong> the line technique, scanning FCS thatprovides millisecond time resolution in a relatively large area. This technique can provide themobile and immobile fraction <strong>of</strong> molecules in a region <strong>of</strong> the cell as well as in<strong>for</strong>mation on localvelocity gradients.13
F<strong>Biological</strong> <strong>Imaging</strong>New Dimensions in In Vivo <strong>Imaging</strong>Lecture AbstractQuantitative Fluorescent Speckle Microscopy in Studies <strong>of</strong> the Cytoskeleton inCell MigrationDr. Clare Waterman-StorerThe Scripps Research InstituteLa Jolla, CAAuditorium2:50-3:30PMluorescent Speckle Microscopy (FSM) is a technology <strong>for</strong> analyzing the dynamics <strong>of</strong>macromolecular assemblies. Originally, the effect <strong>of</strong> random fluorescent speckle <strong>for</strong>mation wasdiscovered with the microtubule cytoskeletal polymer. Since then, the method has been expanded toother proteins <strong>of</strong> the cytoskeleton such as f-actin and microtubule binding proteins. Newlydeveloped, specialized s<strong>of</strong>tware <strong>for</strong> analyzing speckle movement and photometric fluctuation in thecontext <strong>of</strong> polymer transport and turnover has turned FSM into a powerful method <strong>for</strong> the study <strong>of</strong>cytoskeletal dynamics in cell migration, division, morphogenesis, and neuronal path finding. In allthese settings FSM serves as the quantitative readout to link molecular and genetic interventions tocomplete maps <strong>of</strong> the cytoskeleton dynamics and thus can be used <strong>for</strong> the systematic deciphering <strong>of</strong>molecular regulation <strong>of</strong> the cytoskeleton. We envision that FSM has the potential to become a coretool in automated, cell-based molecular diagnostics in cases where variations in cytoskeletaldynamics are a sensitive signal <strong>for</strong> the state <strong>of</strong> a disease, or the activity <strong>of</strong> a molecular perturbant. Iwill review the origins <strong>of</strong> FSM, discuss the most recent technical developments and give a glimpse t<strong>of</strong>uture directions and potentials <strong>of</strong> FSM.14
S<strong>Biological</strong> <strong>Imaging</strong>New Dimensions in In Vivo <strong>Imaging</strong>Lecture AbstractQdot TM Conjugates in <strong>Biological</strong> <strong>Imaging</strong>Dr. Marcel BruchezPrincipal ScientistQuantum Dot CorporationHayward, CAAuditorium3:30-4:10PMemiconductor quantum dots are emerging as a powerful new tool <strong>for</strong> fluorescence detection inbiological systems. As a material, these quantum dots have large absorbance cross-sections,high quantum yields, and very narrow and tunable emission spectra, all <strong>of</strong> which translate to highlysensitive multiplexed detection in biological systems. Using a range <strong>of</strong> sizes <strong>of</strong> CdSe materials, wehave prepared quantum dot probes that span the visible wavelength range from blue-green to red.To date, though there have not been robust, simple chemistries that are available <strong>for</strong> routinemodification <strong>of</strong> the quantum dots to alter their physical, chemical, or biological properties. We havedeveloped new chemistries based on amphiphilic polymers <strong>for</strong> functionalizing these nanoparticlesthat are simple, stable and maintain the underlying optical properties. These materials serve as thestarting materials <strong>for</strong> further refinement and optimization <strong>of</strong> the colloid and biological properties <strong>of</strong>these materials. We have reduced the non-specific binding <strong>of</strong> these materials in cellular assays withboth live and fixed cells by chemically modifying the surface with a variety <strong>of</strong> functional groups.We have used these materials with reduced nonspecific binding to prepare conjugated probes <strong>of</strong>bio-molecules with dramatically improved per<strong>for</strong>mance in cellular imaging and in-vivo imaging <strong>of</strong>isolated cells and in live animals. The per<strong>for</strong>mance advantages <strong>of</strong> these materials in biologicaldetection and multiplexing will be highlighted.15
W<strong>Biological</strong> <strong>Imaging</strong>New Dimensions in In Vivo <strong>Imaging</strong>Lecture AbstractSite-Specific Localization <strong>of</strong> Small Molecule Reporters Within Living CellsDr. Georgyi LosCellular Analysis, Promega CorporationMadison, WIAuditorium4:10-4:50PMith the completion <strong>of</strong> genome sequencing ef<strong>for</strong>ts it is now important to understand how theproducts <strong>of</strong> these genes function, and most importantly how they function within living cellsto dictate the biology <strong>of</strong> organisms. The ability to specifically label proteins can help revealin<strong>for</strong>mation about protein functions and dynamics within the complex biochemical environment <strong>of</strong>the living cells. <strong>Here</strong> we describe a novel technology <strong>for</strong> covalently tethering functional groups[(FG), e.g. fluorescent dye] to a universal reporting protein (URP) within cells. The URP is a mutantenzyme capable <strong>of</strong> <strong>for</strong>ming a stable covalent bond to a modified substrate coupled to the functionalgroup. In our initial approach the URP is a halo-alkane dehalogenase from Rhodococcus rhodochrous(DhaA) with a mutation in a critical active site residue. The activity <strong>of</strong> DhaA cleaves carbon-halogenbonds in aliphatic and aromatic halogenated compounds involving a typical hydrolytic triad. In thisreaction an enzyme-substrate complex is <strong>for</strong>med by a nucleophilic attack involving Asp106 and the<strong>for</strong>mation <strong>of</strong> an ester intermediate; His272 activates H 2 0 that hydrolyzes this intermediate releasingproduct from the catalytic center. A point mutation in DhaA involving a substitution <strong>of</strong>phenylalanine <strong>for</strong> His272 impairs the hydrolysis step leading to a stable covalent intermediate withsubstrate and any conjugated FG. This technology allows the labeling <strong>of</strong> mutant DhaA or mutantDhaA fusion proteins expressed in cells. A significant advantage <strong>of</strong> this approach is the flexibility tocreate labeled proteins with a potentially wide range <strong>of</strong> optical properties or other functionalities.The technology can be applied in different cells and organisms allowing a variety <strong>of</strong> experimentalapproaches to study protein function in living cells.16
T<strong>Biological</strong> <strong>Imaging</strong>New Dimensions in In Vivo <strong>Imaging</strong>Lecture AbstractIn Vivo Two-Photon MicroscopyDr. Wen-Biao GanSkirball Institute, New York <strong>University</strong> School <strong>of</strong> MedicineNew York, NYAuditorium4:50-5:30PMhe invention <strong>of</strong> two-photon microscopy has dramatically enhanced the field <strong>of</strong> laser scanningmicroscopy. Unlike single-photon excitation used in confocal microscopy, two-photonexcitation depends on nearly simultaneous absorption <strong>of</strong> two photons by a single fluorophore. Eachphoton used <strong>for</strong> excitation in two-photon microscopy has approximately half the energy (or twicethe wavelength) <strong>of</strong> the one in confocal microscopy. Due to the use <strong>of</strong> longer wavelength light andthe occurrence <strong>of</strong> two-photon excitation only at the focal point, two-photon microscopy allowsincreased tissue penetration, reduced photo-damage and more efficient fluorescence detection incomparison with confocal microscopy. These advantages are especially important <strong>for</strong> monitoringthick living tissues because multiple exposures <strong>of</strong>ten cause significant photo-toxicity. Asdemonstrated in many recent studies, two-photon microcopy excels in imaging thick biologicalspecimens such as living brains and other intact tissues.We have recently developed a transcranial two-photon imaging technique to study long-termchanges <strong>of</strong> synaptic structures in transgenic mice expressing Yellow Fluorescent Protein (YFP). Byfollowing identified dendritic spines <strong>of</strong> pyramidal neurons in the primary visual cortex, we foundthat the overwhelming majority <strong>of</strong> spines (~96%) remain stable over a one-month interval, with ahalf-life greater than 13 months in adulthood. These results indicate that dendritic spines areremarkably stable in the adult, providing a potential structural basis <strong>for</strong> long-term in<strong>for</strong>mationstorage. In combination with the generation <strong>of</strong> transgenic animals over expressing fluorescentmarkers, in vivo two-photon microscopy opens exciting new avenues <strong>for</strong> studying not only basicbrain function, but also long-term changes seen in neurodegenerative diseases such as Alzheimer’sdisease.17
T<strong>Biological</strong> <strong>Imaging</strong>New Dimensions in In Vivo <strong>Imaging</strong>Keynote Speaker AbstractMulti-Scale Structure and Function in the Nervous SystemDr. Mark EllismanPr<strong>of</strong>essor <strong>of</strong> Neurosciences and BioengineeringDirector, National Center <strong>for</strong> Microscopy and <strong>Imaging</strong> Research<strong>University</strong> <strong>of</strong> Cali<strong>for</strong>nia, San DiegoLa Jolla, CAAuditorium7:30-9:00PMhe activities <strong>of</strong> the National Center <strong>for</strong> Microscopy and <strong>Imaging</strong> Research in San Diego in thedevelopment <strong>of</strong> novel techniques <strong>for</strong> 3 dimensional visualization <strong>of</strong> neuronal structures anddirect identification <strong>of</strong> their protein constituents in situ will be described. This includes a newtechnique <strong>for</strong> correlated 4 dimensional light and 3 dimensional electron microscopy. A continuingchallenge to neuroscientists and biomedical researchers in general is the understanding <strong>of</strong> structureson the scale <strong>of</strong> 1 nm3 to 10's <strong>of</strong> µm3, a dimensional range that encompasses macromolecularcomplexes, organelles, and multi-component structures like synapses and the cellular interactionswithin tissues. On the smaller end <strong>of</strong> this range, structures have traditionally been difficult to studybecause they fall in the resolution gap between technologies, such as X-ray crystallography andelectron microscopy on the lower end, and it is challenging to make correlations between 3D datafrom light and electron microscopy on the upper end. But the continuum <strong>of</strong> in<strong>for</strong>mation will have tobe mapped if the results <strong>of</strong> the molecular revolution, the protein products <strong>of</strong> sequenced genomes,are to be situated in their proper subcellular, cellular and tissue contexts. The technique <strong>of</strong> electrontomography, the derivation <strong>of</strong> 3D structure from a series <strong>of</strong> 2D electron microscopic images, hasgone a long way towards bridging the resolution gaps. While electron tomography is a useful technique <strong>for</strong> providing in<strong>for</strong>mation in this spatial domain, development <strong>of</strong> new methods to provideselective contrast to molecular constituents, organelle complexes, cells and cell processes arerequired. We recently developed a new molecular-tagging technique to chronicle the development,movement and interactions <strong>of</strong> proteins as they do their work in living cells. [Gaietta et al., ScienceApril 19, 2002]. This multi-scale molecular tagging technology involves the genetic tagging <strong>of</strong> aprotein with a small binding area (a tetracystine domain between six to 20 amino-acids long), thatthen interacts with a variety <strong>of</strong> other compounds. Results with two <strong>of</strong> these compounds, greenFlAsH and red ReAsH, have been used in sequence to provide the ability to monitor the trafficking<strong>of</strong> proteins in cells. The red label, ReAsh, can then be used to generate reactive oxygen and drive thedeposition <strong>of</strong> diamino benzadine <strong>for</strong> direct EM-level detection <strong>of</strong> the same protein complex. The use<strong>of</strong> this new labeling method, other multi-scale staining techniques and new advanced imaginginstruments will be described in the context <strong>of</strong> work create a Biomedical In<strong>for</strong>matics ResearchNetwork (BIRN). BIRN is a leading example <strong>of</strong> a virtual distributed database ef<strong>for</strong>t that is federatingmultiscale data about the nervous systems.18
<strong>Biological</strong> <strong>Imaging</strong>New Dimensions in In Vivo <strong>Imaging</strong>VendorsCarl Zeiss, Inc., LSM 5 Pascal Microscope (Room 215)The LSM 5 PASCAL is a confocal laser scanning microscope <strong>for</strong> basic research in medicine and biology.Equipped with optimized, user friendly hardware and s<strong>of</strong>tware, this system delivers excellent two– andthree-dimensional images, especially in fluorescence applications. It is ideal <strong>for</strong> the acquisition <strong>of</strong> time series inphysiological experiments as well as <strong>for</strong> quantitative measurements.Leica Microsystems, TCS SP2 AOBS Microscope (Room 122)The Leica TCS SP2 AOBS integrates multiple innovations <strong>of</strong> Leica Microsystems R & D into a research systemthat sets new standards <strong>for</strong> efficient protection <strong>of</strong> live cells and flexibility. Designed as a system in harmony itbegins with special "confocal" objectives, versatile microscope stands and diffraction limited optics;preconditions <strong>for</strong> outstanding image quality and a must <strong>for</strong> meaningful results in live cell research. Milestonesin optical innovation include our multi-laser merge module with programmable control <strong>of</strong> excitationwavelengths and intensity, beam scanning with our proprietary K scanner and our highly efficient andselective SP prism spectrophotometer <strong>for</strong> detection <strong>of</strong> fluorescence emission.Mad City Labs (Room 122)Mad City Labs, Inc is the leading American manufacturer <strong>of</strong> nanopositioning systems with sub-nanometerprecision at an af<strong>for</strong>dable price. We have the largest product line <strong>of</strong> nanopositioning systems available andspecialize in custom nanopositioner applications. We deliver the tools <strong>for</strong> nanotechnology in 30 to 45 days, andprovide the highest level <strong>of</strong> customer service and satisfaction in the industry. Applications <strong>for</strong> nanopositionersinclude AFM, NSOM, Scanned Probe Microscopy, fiber positioning, interferometry, single moleculespectroscopy and lithography.Nikon C1 Confocal Microscope System (Room 122)A universal confocal microscope system that is ultra-compact and lightweight yet provides confocal images <strong>of</strong>the highest quality in its class. Highest optical per<strong>for</strong>mance; 4-channel simultaneous detection; Four positionpinhole turret; 12 bit imaging; Optical fiber coupling; Intuitive s<strong>of</strong>tware; Advanced acquisition and analysisfeatures; 50 nm steps along the Z axis; CF160 optical system; Advanced accessories and various lasercombinations are available.The Fryer Company, (Room 122) located in Huntley, Illinois, is one <strong>of</strong> the largest distributors <strong>of</strong> NikonBiomedical and Industrial microscopes in the United States. Over the past 40 years, Fryer has become a leading<strong>for</strong>ce in marketing innovative imaging products throughout the medical, industrial and research fields.19
<strong>Biological</strong> <strong>Imaging</strong>New Dimensions in In Vivo <strong>Imaging</strong>SponsorsBio-Rad Laboratories, Inc. is a multinational manufacturer and distributor <strong>of</strong> life scienceresearch products and clinical diagnostics. Its Cell Science Division manufactures a range <strong>of</strong> laser-microscopeproducts. These include confocal and multi-photon laser scanning imaging systems (Radiance2100) and cellisolation systems (Clonis). The latest version <strong>of</strong> the Radiance confocal imaging system - the 'Radiance Rainbow'adds spectral discrimination and the ability to reassign (unmix) the emission from overlapping fluorophores.20
Bio-Rad Laboratories—microscopy.bio-rad.comCarl Zeiss, Inc.—zeiss.comLeica Microsystems—leica-microscopy.comLOCI Group—loci.wisc.eduMad City Labs —madcitylabs.com<strong>Biological</strong> <strong>Imaging</strong>New Dimensions in In Vivo <strong>Imaging</strong>Websites Associates with this SymposiumNational Center <strong>for</strong> Microscopy and <strong>Imaging</strong> Research—ncmir.ucsd.eduNew York <strong>University</strong> School <strong>of</strong> Medicine—med.nyu.eduNikon—microscopyU.comPromega Corporation—promega.comPromega R&D—promega-rd.infoQuantum Dot Corporation—qdots.comScripps Research Institute—scripps.eduThe Fryer Company—fryerco.comUniversal <strong>Imaging</strong>—image1.com<strong>University</strong> <strong>of</strong> Connecticut Health Center—uchc.edu<strong>University</strong> <strong>of</strong> Illinois at Urbana-Champaign—uiuc.edu<strong>University</strong> <strong>of</strong> Wisconsin-Madison—wisc.eduW.M. <strong>Keck</strong> <strong>Laboratory</strong>—keck.bioimaging.wisc.eduWellman <strong>Laboratory</strong> <strong>of</strong> Photomedicine—mgh.harvard.edu/wellmanSecond Symposium on <strong>Biological</strong> <strong>Imaging</strong>—promega-rd.info/bioimage2003/default.asp21
<strong>Biological</strong> <strong>Imaging</strong>New Dimensions in In Vivo <strong>Imaging</strong>Notes22
<strong>Biological</strong> <strong>Imaging</strong>New Dimensions in In Vivo <strong>Imaging</strong>Notes23
<strong>Biological</strong> <strong>Imaging</strong>New Dimensions in In Vivo <strong>Imaging</strong>Notes24