Probabilistic Model Checking for Systems Biology - PRISM
Probabilistic Model Checking for Systems Biology - PRISM
Probabilistic Model Checking for Systems Biology - PRISM
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
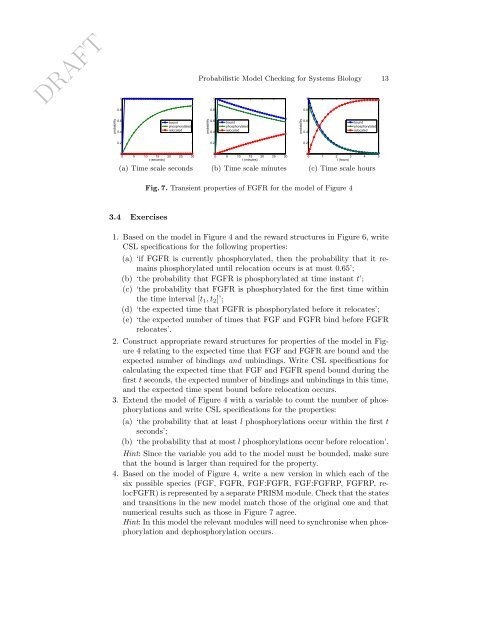
DRAFT10.8<strong>Probabilistic</strong> <strong>Model</strong> <strong>Checking</strong> <strong>for</strong> <strong>Systems</strong> <strong>Biology</strong> 13110.80.8probability0.60.4boundphosphorylatedrelocatedprobability0.60.4boundphosphorylatedrelocatedprobability0.60.4boundphosphorylatedrelocated0.20.20.200 5 10 15 20 25 30t (seconds)(a) Time scale seconds00 5 10 15 20 25 30t (minutes)(b) Time scale minutes00 1 2 3 4 5t (hours)(c) Time scale hoursFig. 7. Transient properties of FGFR <strong>for</strong> the model of Figure 43.4 Exercises1. Based on the model in Figure 4 and the reward structures in Figure 6, writeCSL specifications <strong>for</strong> the following properties:(a) ‘if FGFR is currently phosphorylated, then the probability that it remainsphosphorylated until relocation occurs is at most 0.65’;(b) ‘the probability that FGFR is phosphorylated at time instant t’;(c) ‘the probability that FGFR is phosphorylated <strong>for</strong> the first time withinthe time interval [t 1 , t 2 ]’;(d) ‘the expected time that FGFR is phosphorylated be<strong>for</strong>e it relocates’;(e) ‘the expected number of times that FGF and FGFR bind be<strong>for</strong>e FGFRrelocates’.2. Construct appropriate reward structures <strong>for</strong> properties of the model in Figure4 relating to the expected time that FGF and FGFR are bound and theexpected number of bindings and unbindings. Write CSL specifications <strong>for</strong>calculating the expected time that FGF and FGFR spend bound during thefirst t seconds, the expected number of bindings and unbindings in this time,and the expected time spent bound be<strong>for</strong>e relocation occurs.3. Extend the model of Figure 4 with a variable to count the number of phosphorylationsand write CSL specifications <strong>for</strong> the properties:(a) ‘the probability that at least l phosphorylations occur within the first tseconds’;(b) ‘the probability that at most l phosphorylations occur be<strong>for</strong>e relocation’.Hint: Since the variable you add to the model must be bounded, make surethat the bound is larger than required <strong>for</strong> the property.4. Based on the model of Figure 4, write a new version in which each of thesix possible species (FGF, FGFR, FGF:FGFR, FGF:FGFRP, FGFRP, relocFGFR)is represented by a separate <strong>PRISM</strong> module. Check that the statesand transitions in the new model match those of the original one and thatnumerical results such as those in Figure 7 agree.Hint: In this model the relevant modules will need to synchronise when phosphorylationand dephosphorylation occurs.