Molecular phylogeny of large miliolid foraminifera - University of ...
Molecular phylogeny of large miliolid foraminifera - University of ...
Molecular phylogeny of large miliolid foraminifera - University of ...
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
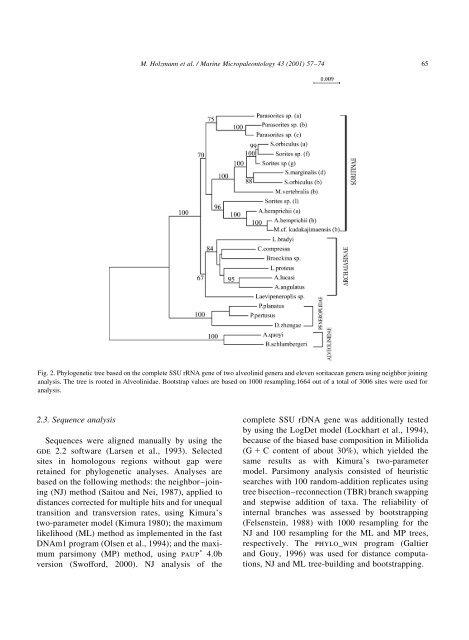
M. Holzmann et al. / Marine Micropaleontology 43 2001) 57±74 65Fig. 2. Phylogenetic tree based on the complete SSU rRNA gene <strong>of</strong> two alveolinid genera and eleven soritacean genera using neighbor joininganalysis. The tree is rooted in Alveolinidae. Bootstrap values are based on 1000 resampling.1664 out <strong>of</strong> a total <strong>of</strong> 3006 sites were used foranalysis.2.3. Sequence analysisSequences were aligned manually by using thegde 2.2 s<strong>of</strong>tware Larsen et al., 1993). Selectedsites in homologous regions without gap wereretained for phylogenetic analyses. Analyses arebased on the following methods: the neighbor±joiningNJ) method Saitou and Nei, 1987), applied todistances corrected for multiple hits and for unequaltransition and transversion rates, using Kimura'stwo-parameter model Kimura 1980); the maximumlikelihood ML) method as implemented in the fastDNAm1 program Olsen et al., 1994); and the maximumparsimony MP) method, using paup p 4.0bversion Sw<strong>of</strong>ford, 2000). NJanalysis <strong>of</strong> thecomplete SSU rDNA gene was additionally testedby using the LogDet model Lockhart et al., 1994),because <strong>of</strong> the biased base composition in MiliolidaG 1 C content <strong>of</strong> about 30%), which yielded thesame results as with Kimura's two-parametermodel. Parsimony analysis consisted <strong>of</strong> heuristicsearches with 100 random-addition replicates usingtree bisection±reconnection TBR) branch swappingand stepwise addition <strong>of</strong> taxa. The reliability <strong>of</strong>internal branches was assessed by bootstrappingFelsenstein, 1988) with 1000 resampling for theNJand 100 resampling for the ML and MP trees,respectively. The phylo_win program Galtierand Gouy, 1996) was used for distance computations,NJand ML tree-building and bootstrapping.