Analysis of Amino Acids Contained in a Grain of Rice - Shimadzu
Analysis of Amino Acids Contained in a Grain of Rice - Shimadzu
Analysis of Amino Acids Contained in a Grain of Rice - Shimadzu
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
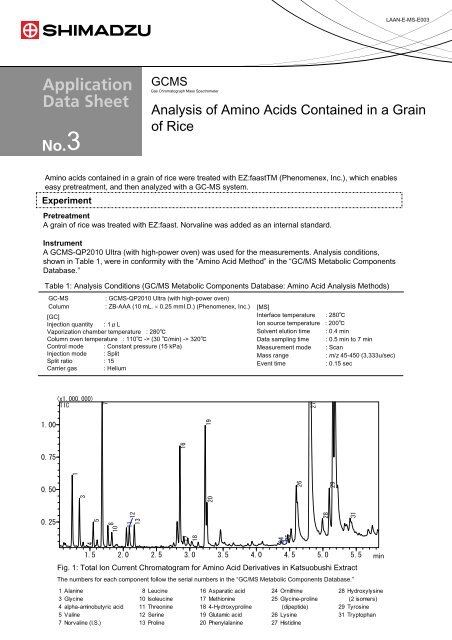
LAAN-E-MS-E003GCMSGas Chromatograph Mass Spectrometer3<strong>Analysis</strong> <strong>of</strong> <strong>Am<strong>in</strong>o</strong> <strong>Acids</strong> <strong>Conta<strong>in</strong>ed</strong> <strong>in</strong> a Gra<strong>in</strong><strong>of</strong> <strong>Rice</strong><strong>Am<strong>in</strong>o</strong> acids conta<strong>in</strong>ed <strong>in</strong> a gra<strong>in</strong> <strong>of</strong> rice were treated with EZ:faastTM (Phenomenex, Inc.), which enableseasy pretreatment, and then analyzed with a GC-MS system.ExperimentPretreatmentA gra<strong>in</strong> <strong>of</strong> rice was treated with EZ:faast. Norval<strong>in</strong>e was added as an <strong>in</strong>ternal standard.InstrumentA GCMS-QP2010 Ultra (with high-power oven) was used for the measurements. <strong>Analysis</strong> conditions,shown <strong>in</strong> Table 1, were <strong>in</strong> conformity with the “<strong>Am<strong>in</strong>o</strong> Acid Method” <strong>in</strong> the “GC/MS Metabolic ComponentsDatabase.”Table 1: <strong>Analysis</strong> Conditions (GC/MS Metabolic Components Database: <strong>Am<strong>in</strong>o</strong> Acid <strong>Analysis</strong> Methods)GC-MS: GCMS-QP2010 Ultra (with high-power oven)Column : ZB-AAA (10 mL. × 0.25 mmI.D.) (Phenomenex, Inc.) [MS][GC]Interface temperature : 280℃Injection quantity : 1μLIon source temperature : 200℃Vaporization chamber temperature : 280℃Solvent elution time : 0.4 m<strong>in</strong>Column oven temperature : 110℃ -> (30 ℃/m<strong>in</strong>) -> 320℃Data sampl<strong>in</strong>g time : 0.5 m<strong>in</strong> to 7 m<strong>in</strong>Control mode : Constant pressure (15 kPa)Measurement mode : ScanInjection mode : SplitMass range: m/z 45-450 (3,333u/sec)Split ratio : 15Event time: 0.15 secCarrier gas : Helium(x1,000,000)TIC1.000.7510.5030.2547581011 12131718161924252027262928311.5 2.0 2.5 3.0 3.5 4.0 4.5 5.0 5.5Fig. 1: Total Ion Current Chromatogram for <strong>Am<strong>in</strong>o</strong> Acid Derivatives <strong>in</strong> Katsuobushi ExtractThe numbers for each component follow the serial numbers <strong>in</strong> the “GC/MS Metabolic Components Database.”1 Alan<strong>in</strong>e 8 Leuc<strong>in</strong>e 16 Asparatic acid 24 Ornith<strong>in</strong>e 28 Hydroxylys<strong>in</strong>e3 Glyc<strong>in</strong>e 10 Isoleuc<strong>in</strong>e 17 Methion<strong>in</strong>e 25 Glyc<strong>in</strong>e-prol<strong>in</strong>e (2 isomers)4 alpha-am<strong>in</strong>obutyric acid 11 Threon<strong>in</strong>e 18 4-Hydroxyprol<strong>in</strong>e (dipeptide) 29 Tyros<strong>in</strong>e5 Val<strong>in</strong>e 12 Ser<strong>in</strong>e 19 Glutamic acid 26 Lys<strong>in</strong>e 31 Tryptophan7 Norval<strong>in</strong>e (I.S.) 13 Prol<strong>in</strong>e 20 Phenylalan<strong>in</strong>e 27 Histid<strong>in</strong>em<strong>in</strong>
33.0 (x100,000)TIC2.52.01.51.00.5134578101112 13141619201.5 2.0 2.5 3.0 3.5 4.0 4.5 5.0 5.5 6.0m<strong>in</strong>Fig. 2: Total Ion Current Chromatogram for <strong>Am<strong>in</strong>o</strong> Acid Derivatives <strong>in</strong> Kombu ExtractThe numbers for each component follow the serial numbers <strong>in</strong> the “GC/MS Metabolic Components Database.”23262931321 Alan<strong>in</strong>e 8 Leuc<strong>in</strong>e 14 Asparag<strong>in</strong>e 26 Lys<strong>in</strong>e3 Glyc<strong>in</strong>e 10 Isoleuc<strong>in</strong>e 16 Aspartic acid 29 Tyros<strong>in</strong>e4 alpha-am<strong>in</strong>obutyric acid 11 Threon<strong>in</strong>e 19 Glutamic acid 31 Tryptophan5 Val<strong>in</strong>e 12 Ser<strong>in</strong>e 20 Phenylalan<strong>in</strong>e 32 Cystathion<strong>in</strong>e7 Norval<strong>in</strong>e(I.S.) 13 Prol<strong>in</strong>e 23 Glutam<strong>in</strong>eSummaryPretreatment us<strong>in</strong>g the EZ:faast kit, followed by analysis us<strong>in</strong>g the GCMS-QP2010 Ultra, which isequipped with a high-speed scann<strong>in</strong>g function, enabled rapid analysis <strong>of</strong> am<strong>in</strong>o acids. With thiscomb<strong>in</strong>ation, it took only 15 m<strong>in</strong>utes per sample from pretreatment to analysis.(Reference: <strong>Shimadzu</strong> Application News No. M246 <strong>Analysis</strong> <strong>of</strong> <strong>Am<strong>in</strong>o</strong> <strong>Acids</strong> Us<strong>in</strong>g Fast-GC/MS andMetabolite Database)For Research Use Only. Not for use <strong>in</strong> diagnostic procedures.<strong>Shimadzu</strong> Corporation (“<strong>Shimadzu</strong>”) reserves all rights <strong>in</strong>clud<strong>in</strong>g copyright <strong>in</strong> this publication. <strong>Shimadzu</strong> does not assume any responsibility or liability for any damage, whether direct or<strong>in</strong>direct, relat<strong>in</strong>g to, or aris<strong>in</strong>g out <strong>of</strong> the use <strong>of</strong> this publication. This publication is based upon the <strong>in</strong>formation available to <strong>Shimadzu</strong> on or before the date <strong>of</strong> publication, and subject tochange without notice.First Edition: September 2011