RNase Undetectable In Water After Ultrafiltration
RNase Undetectable In Water After Ultrafiltration
RNase Undetectable In Water After Ultrafiltration
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
26 [Tools & Techniques] www.biosciencetechnology.com<br />
I<br />
<strong>Ultrafiltration</strong><br />
removes <strong>RNase</strong><br />
from water to below<br />
detectable<br />
limits.<br />
<strong>RNase</strong> <strong>Undetectable</strong> <strong>In</strong> <strong>Water</strong><br />
<strong>After</strong> <strong>Ultrafiltration</strong><br />
Joseph Plurad and Stéphane Mabic<br />
<strong>In</strong>troduction<br />
Highly purified water has become a standard tool in molecular<br />
biology. Techniques such as reverse osmosis, electrodeionization,<br />
ultrafiltration, and chemical treatments provide water of varying<br />
quality and utility. <strong>Water</strong> quality specifications differ for applications<br />
such as liquid chromatography, in situ hybridizations, single<br />
nucleotide polymorphism analysis, and DNA amplification.<br />
Experiments involving reverse-transcriptase PCR (RT-PCR), plasmid<br />
preparation, construction of cDNA libraries, and Northern blotting<br />
minimally require <strong>RNase</strong>-free water. However, RNA preparations<br />
from cells and tissues are complicated by the presence of<br />
ribonucleases (<strong>RNase</strong>s).<br />
<strong>Water</strong> treatment<br />
Autoclaving and heat treatment do not satisfactorily inactivate<br />
<strong>RNase</strong>, which can survive being heated to 180 C for 4<br />
hours. By killing bacteria, autoclaving promotes release<br />
of bacterial <strong>RNase</strong>s, which may regain significant<br />
activity at experimental temperatures.<br />
Treatment with diethylpyrocarbonate (DEPC),<br />
an irreversible <strong>RNase</strong> inhibitor, has evolved as<br />
the standard method for preparing <strong>RNase</strong>-free<br />
water for PCR. DEPC reacts with an active-site<br />
histidine residue on <strong>RNase</strong>, forming a carbamate<br />
and liberating a molecule of carbon dioxide and<br />
ethanol. Since the chemical reaction is irreversible,<br />
<strong>RNase</strong> inactivation by DEPC is presumed to be complete<br />
and permanent. Although vendors and end-users<br />
employ DEPC to produce PCR-quality nuclease-free water, the<br />
DEPC method suffers from several drawbacks.<br />
As a chemical treatment, DEPC merely inactivates <strong>RNase</strong> without<br />
removing it. <strong>In</strong>activated enzyme remains in solution. <strong>After</strong> a onehour<br />
incubation, excess DEPC is destroyed by being heated to 121 C,<br />
hydrolyzing pyrocarbonate to carbon dioxide and generating ethanol<br />
as the side-product. Most of the ethanol evaporates from solution.<br />
What remains of side products and spent reagent contributes significantly<br />
to the water’s total organic carbon (TOC) content.<br />
By contributing carbonate/bicarbonate, DEPC decreases resistivity<br />
and lowers the solution’s pH, which may contribute to RNA<br />
instability. As we have observed by monitoring DEPC-treated<br />
water, inactivated <strong>RNase</strong> may regain some activity over time.<br />
TOC content and conductivity value of various purified waters<br />
from Millipore illustrates the potential unsuitability of DEPC-treated<br />
water for sensitive experiments. DEPC-treated water from a<br />
major vendor contained a TOC of 122,800 ppb and exhibited conductivity<br />
of 2.9 µS/cm. Milli-Q water treated with DEPC was<br />
somewhat better, with a TOC of 15,700 ppb, but even higher conductivity<br />
than commercial DEPC-treated water, 3.4 µS/cm.<br />
Although two other commercial <strong>RNase</strong>-free waters contained much<br />
lower levels of TOC (821 and 361 ppb), they showed high conductivities<br />
of 2.1 and 2.8 µS/cm, respectively.<br />
<strong>Ultrafiltration</strong> (UF), the non-chemical alternative to DEPC treatment,<br />
removes <strong>RNase</strong> from water to below detectable limits and<br />
thereby removes, rather than contributing to, TOC. Typical UF<br />
methods employ polysulfone UF membranes embedded in acrylonitrile<br />
butadiene styrene polymer (ABS) housings. Neither membrane<br />
nor housing materials contribute to product water’s ion or organic<br />
content. Milli-Q water treated by ultrafiltration contained less than<br />
15 ppb of TOC and conductivity of 0.055 µS/cm corresponding to<br />
18.2 megohm cm resistivity.<br />
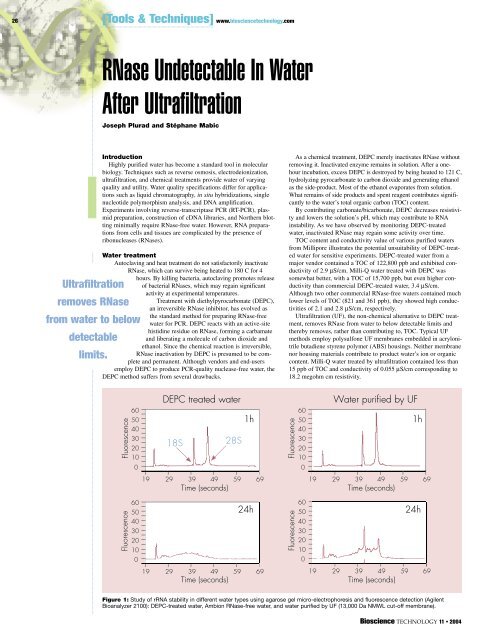
Figure 1: Study of rRNA stability in different water types using agarose gel micro-electrophoresis and fluorescence detection (Agilent<br />
Bioanalyzer 2100): DEPC-treated water, Ambion <strong>RNase</strong>-free water, and water purified by UF (13,000 Da NMWL cut-off membrane).<br />
Bioscience TECHNOLOGY 11 • 2004
28 [Tools & Techniques] www.biosciencetechnology.com<br />
Examples<br />
Quantitative <strong>RNase</strong> recovery at high flow<br />
<strong>In</strong> one experiment, a polysulfone<br />
Millipore Pyrogard UF membrane with a<br />
nominal molecular weight cutoff of<br />
13,000 Da was challenged with 200 liter<br />
solutions containing 0.1 ng/ml of <strong>RNase</strong>A<br />
(Roche Diagnostics, 109 142) at a<br />
throughput of 500 ml/min. This concentration<br />
of <strong>RNase</strong> is between 10-100 fold<br />
various water samples as measured by the<br />
<strong>RNase</strong>Alert assay. The table suggests that<br />
<strong>RNase</strong> is ubiquitous in both treated and<br />
untreated water samples. Even highly<br />
purified water (reverse osmosis, Elix<br />
water systems, unattended Milli-Q<br />
Gradient systems) contains significant<br />
<strong>RNase</strong> activity that could affect PCR<br />
results. As expected, <strong>RNase</strong>-free, DEPCtreated<br />
waters, and all UF-treated samples<br />
Figure 2: Quantitative RT-PCR: Correlation between theoretical and experimental data.<br />
higher than is normally encountered in most<br />
PCR experiments. Total weight of collected<br />
<strong>RNase</strong> A was 22 mg, which was essentially<br />
quantitative (200 liters � 0.1 ng/ml).<br />
<strong>RNase</strong> in sample waters<br />
<strong>RNase</strong> activity was measured at various<br />
stages of water purification with the<br />
<strong>RNase</strong>Alert test kit (Ambion), which<br />
claims a lower detection limit of 0.003<br />
ng/ml. <strong>RNase</strong>Alert uses a cleavable,<br />
<strong>RNase</strong> substrate standard labeled with a<br />
green fluorescent probe. <strong>After</strong> generating<br />
a calibration curve using an <strong>RNase</strong> standard<br />
(Ambion), we tested samples on an<br />
SFM 25 Konitron fluorometer, excited at<br />
520 nm and read at 490 nm.<br />
Table 1 lists <strong>RNase</strong> concentrations in<br />
(Millipore Pyrogard D UF cartridge;<br />
13,000 Da NMWL cutoff) showed the<br />
lowest <strong>RNase</strong> levels,