EMS Newsletter June 2010 - European Mathematical Society ...
EMS Newsletter June 2010 - European Mathematical Society ...
EMS Newsletter June 2010 - European Mathematical Society ...
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
quence, close to it (i.e. a few mutations away) there will be a<br />
“cloud” of less adapted sequences coexisting with the master<br />
sequence. Quasispecies is the term that Eigen introduced to<br />
describe this ensemble of sequences.<br />
In order to study the behaviour of a quasispecies more<br />
closely we will resort to equation (9). For simplicity we shall<br />
assume that sequences have finite length L � 1, that there<br />
are just two alleles per locus, that the master sequence has<br />
fitness f1 = f > 1 and that all other sequences have fitness<br />
f2 = ··· = f 2 L = 1. We shall also assume that the probability<br />
that a point mutation occurs is μ � 1, independent of the<br />
sequence. Let x1 = x denote the population fraction of the<br />
master sequence; thus x2 +···+ x 2 L = 1− x and φ = fx+1− x.<br />
Equation (9) then becomes<br />
˙x = x � f (1 − μ) L − 1 − ( f − 1)x � + O(μ). (34)<br />
The term O(μ) accounts for the transitions from the L nearest<br />
neighbour sequences of the master sequence that revert<br />
to the master sequence. Neglecting these terms and approximating<br />
(1 − μ) L ≈ e −Lμ we can see that if fe −Lμ > 1 then x<br />
asymptotically approaches x ∗ = (e −Lμ f − 1)/( f − 1), whereas<br />
if fe −Lμ < 1 the bracket in equation (34) becomes negative<br />
and therefore x = O(μ). The threshold μerr = log f /L defines<br />
the error catastrophe. When μμerr the identity of this master sequence<br />
gets lost in the cloud of mutants and the quasispecies disappears<br />
as such.<br />
Experimental studies performed in the ’90s seem to confirm<br />
[16] that indeed the length of the genome of different<br />
species – ranging from virus to Homo sapiens – and the mutation<br />
rate per base are related as μL ≤ O(1). Hence an increase<br />
in the mutation rate is a mechanism that this theory puts forward<br />
to fight viral infections. We will come back to this point<br />
later.<br />
Rugged landscape<br />
Although locally the adaptive landscape can be well described<br />
by the Fujiyama model, Wright visualized it as a rugged landscape,<br />
full of high peaks separated by deep valleys. The reason<br />
is that mutations that change the sequence minimally may<br />
induce large variations in the fitness of individuals. In addition,<br />
there exists the well known phenomenon of epistasis,<br />
according to which some genes interact, either constructively<br />
or destructively, amplifying these large variations in response<br />
to small changes in the sequences.<br />
According to the rugged landscape metaphor, species evolve<br />
by climbing peaks and sitting on the summits. Different peaks<br />
correspond to different species with different fitnesses. This<br />
picture seems to fit well with our idea of evolution by natural<br />
selection. However, it has a serious drawback: species<br />
that are at a summit can only move to a higher one by going<br />
through an unfit valley. In the most favourable case this valley<br />
will consist of a single intermediate state. Formula (31) tells<br />
us that if a population is small, it is not impossible that an<br />
unfit allele replaces a fitter one. Nevertheless, the probability<br />
that this happens is very small, i.e. adaptation times should<br />
be very large. And this is only the most favourable case. The<br />
high speed of adaptation to rapidly changing environments<br />
Feature<br />
that viruses exhibit seriously challenges this model. What is<br />
then wrong in our picture of adaptive landscapes?<br />
Holey landscape: neutral networks<br />
Let us review the most extreme case of a rugged landscape:<br />
the random landscape. In this case every sequence of X has a<br />
random fitness, independent of the other sequences. In general,<br />
rugged landscapes are not that extreme because there<br />
is some degree of correlation between the fitness of neighbouring<br />
sequences. However, beyond the correlation length,<br />
fitness values become uncorrelated. The random landscape is<br />
the extreme case in which the correlation length is smaller<br />
than 1. Suppose now that the length of the sequences is large,<br />
and that every locus can host A independent alleles. The degree<br />
of graph G will thus be g = (A−1)L and its size |X| = AL .<br />
With L = 100 and A = 2 (a rather modest choice), g = 100<br />
and |X| = 2100 ≈ 1030 . To all purposes such a graph can be<br />
locally approximated by a tree, the more so the larger the degree<br />
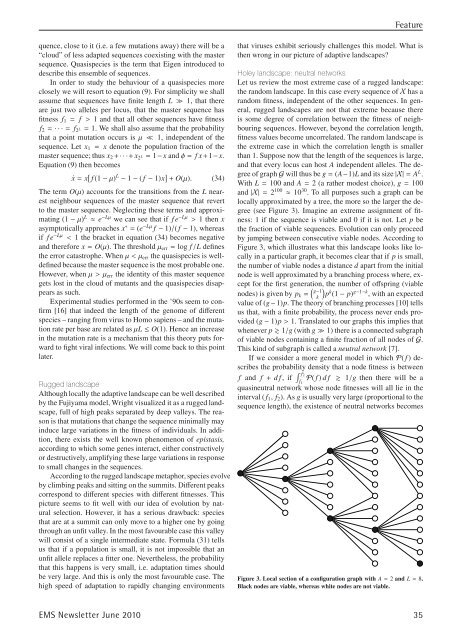
(see Figure 3). Imagine an extreme assignment of fitness:<br />
1 if the sequence is viable and 0 if it is not. Let p be<br />
the fraction of viable sequences. Evolution can only proceed<br />
by jumping between consecutive viable nodes. According to<br />
Figure 3, which illustrates what this landscape looks like locally<br />
in a particular graph, it becomes clear that if p is small,<br />
the number of viable nodes a distance d apart from the initial<br />
node is well approximated by a branching process where, except<br />
for the first generation, the number of offspring (viable<br />
nodes) is given by pk = � �<br />
g−1 k g−1−k<br />
k p (1 − p) , with an expected<br />
value of (g −1)p. The theory of branching processes [10] tells<br />
us that, with a finite probability, the process never ends provided<br />
(g − 1)p > 1. Translated to our graphs this implies that<br />
whenever p � 1/g (with g � 1) there is a connected subgraph<br />
of viable nodes containing a finite fraction of all nodes of G.<br />
This kind of subgraph is called a neutral network [7].<br />
If we consider a more general model in which P( f ) describes<br />
the probability density that a node fitness is between<br />
f and f + df, if � f2<br />
P( f ) df � 1/g then there will be a<br />
f1<br />
quasineutral network whose node fitnesses will all lie in the<br />
interval ( f1, f2). As g is usually very large (proportional to the<br />
sequence length), the existence of neutral networks becomes<br />
Figure 3. Local section of a configuration graph with A = 2 and L = 8.<br />
Black nodes are viable, whereas white nodes are not viable.<br />
<strong>EMS</strong> <strong>Newsletter</strong> <strong>June</strong> <strong>2010</strong> 35