Biochemistry II: Binding of ligands to a macromolecule (or the secret ...
Biochemistry II: Binding of ligands to a macromolecule (or the secret ...
Biochemistry II: Binding of ligands to a macromolecule (or the secret ...
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
<strong>Biochemistry</strong> <strong>II</strong>: <strong>Binding</strong> <strong>of</strong> <strong>ligands</strong> <strong>to</strong> a <strong>macromolecule</strong><br />
(<strong>or</strong> <strong>the</strong> <strong>secret</strong> <strong>of</strong> life itself...)<br />
• http://www.kip.uni-heidelberg.de/chromcon/publications/pdf-files/Rippe_Futura_97.pdf<br />
• Principles <strong>of</strong> Physical <strong>Biochemistry</strong>, van Holde, Johnson & Ho, 1998.<br />
• Slides available at<br />
• http://www.kip.uni-heidelberg.de/chromcon/teaching/index_teaching.html<br />
Karsten Rippe<br />
Kirch<strong>of</strong>f-Institut für Physik<br />
Molecular Biophysics Group<br />
Im Neuenheimer Feld 227<br />
Tel: 54-9270<br />
e-mail: Karsten.Rippe@kip.uni-heidelberg.de
The <strong>secret</strong> <strong>of</strong> life<br />
“The <strong>secret</strong> <strong>of</strong> life is molecular recognition; <strong>the</strong><br />
ability <strong>of</strong> one molecule <strong>to</strong> "recognize" ano<strong>the</strong>r through<br />
weak bonding interactions.”<br />
Linus Pauling at <strong>the</strong> 25th anniversary <strong>of</strong> <strong>the</strong> Institute <strong>of</strong><br />
Molecular Biology at <strong>the</strong> University <strong>of</strong> Oregon
<strong>Binding</strong> <strong>of</strong> dioxygen <strong>to</strong> hemoglobin<br />
(air)
<strong>Binding</strong> <strong>of</strong> Glycerol-Phosphate <strong>to</strong> triose phosphate isomerase<br />
(energy)
Complex <strong>of</strong> <strong>the</strong> HIV protease <strong>the</strong> inhibi<strong>to</strong>r SD146<br />
(drugs)
Antibody HyHEL-10 in complex with Hen Egg White Lyzoyme<br />
(immune system)
X-ray crystal structure <strong>of</strong> <strong>the</strong> TBP-promoter DNA complex<br />
(transcription starts here...)<br />
Nikolov, D. B. & Burley, S. K.<br />
(1997). RNA polymerase <strong>II</strong><br />
transcription initiation: a<br />
structural view. PNAS 94, 15-<br />
22.
Structure <strong>of</strong> <strong>the</strong> tryp<strong>to</strong>phan repress<strong>or</strong> with DNA<br />
(transcription regulation)
<strong>Binding</strong> <strong>of</strong> <strong>ligands</strong> <strong>to</strong> a <strong>macromolecule</strong><br />
• General description <strong>of</strong> ligand binding<br />
– <strong>the</strong> esssentials<br />
– <strong>the</strong>rmodynamics<br />
– Adair equation<br />
• Simple equilibrium binding<br />
– s<strong>to</strong>ichiometric titration<br />
– equilibrium binding/dissociation constant<br />
• Complex equilibrium binding<br />
– cooperativity<br />
– Scatchard plot and Hill Plot<br />
– MWC and KNF model f<strong>or</strong> cooperative binding
The mass equation law f<strong>or</strong> binding <strong>of</strong> a protein P <strong>to</strong> its DNA D<br />
Dfree + P !<br />
free"DP<br />
K1 = Dfree #Pfree DP<br />
binding <strong>of</strong> <strong>the</strong> first proteins with <strong>the</strong> dissociation constant K 1<br />
D free, concentration free DNA; P free, concentration free protein<br />
1<br />
binding constant KB =<br />
dissociation constant KD
What is <strong>the</strong> meaning <strong>of</strong> <strong>the</strong> dissociation constant f<strong>or</strong><br />
binding <strong>of</strong> a single ligand <strong>to</strong> its site?<br />
1. K D is a concentration and has units <strong>of</strong> mol per liter<br />
2. K D gives <strong>the</strong> concentration <strong>of</strong> ligand that saturates 50% <strong>of</strong> <strong>the</strong><br />
sites (when <strong>the</strong> <strong>to</strong>tal sit concentration ismuch lower than K D)<br />
3. Almost all binding sites are saturated if <strong>the</strong> ligand<br />
concentration is 10 x K D<br />
4. The dissociatin constant K D is related <strong>to</strong> Gibbs free energy<br />
∆G by <strong>the</strong> relation ∆G = - R T ln(K D)
K D values in biological systems<br />
Movovalent ions binding <strong>to</strong> proteins <strong>or</strong> DNA have K D 0.1 mM <strong>to</strong> 10 mM<br />
Allosteric activa<strong>to</strong>rs <strong>of</strong> enzymes e. g. NAD have K D 0.1 µM <strong>to</strong> 0.1 mM<br />
Site specific binding <strong>to</strong> DNA K D 1 nM <strong>to</strong> 1 pM<br />
Trypsin inhibi<strong>to</strong>r <strong>to</strong> pancreatic trypsin protease K D 0.01 pM<br />
Antibody-antigen interaction have K D 0.1 mM <strong>to</strong> 0.0001 pM
What is ∆G? The <strong>the</strong>rmodynamics <strong>of</strong> a system<br />
• Biological systems can be usually described as having constant pressure P and constant<br />
temperature T<br />
– <strong>the</strong> system is free <strong>to</strong> exchange heat with <strong>the</strong> surrounding <strong>to</strong> remain at a constant temperature<br />
– it can expand <strong>or</strong> contract in volume <strong>to</strong> remain at atmospheric pressure
Some fundamentals <strong>of</strong> solution <strong>the</strong>rmodynamics<br />
• At constant pressure P and constant temperature T <strong>the</strong> system is described by <strong>the</strong> Gibbs<br />
free energy:<br />
G ! H "T S<br />
• H is <strong>the</strong> enthalpy <strong>or</strong> heat content <strong>of</strong> <strong>the</strong> system, S is <strong>the</strong> entropy <strong>of</strong> <strong>the</strong> system<br />
• a reaction occurs spontaneously only if ∆G < 0<br />
• at equilibrium ∆G = 0<br />
!G = !H "T !S<br />
• f<strong>or</strong> ∆G > 0 <strong>the</strong> input <strong>of</strong> energy is required <strong>to</strong> drive <strong>the</strong> reaction
<strong>the</strong> problem<br />
In general we can not assume that <strong>the</strong> <strong>to</strong>tal free energy G <strong>of</strong> a<br />
solution consisting <strong>of</strong> N different components is simply <strong>the</strong> sum <strong>of</strong> <strong>the</strong><br />
free energys <strong>of</strong> <strong>the</strong> single components.
The chemical potential µ <strong>of</strong> a substance is<br />
<strong>the</strong> partial molar Gibbs free energy<br />
G i = !G<br />
!n i<br />
f<strong>or</strong> an ideal solution it is:<br />
n<br />
= µ G = " n i i!µ i<br />
i=1<br />
µ i = µ i 0 f<strong>or</strong> Ci =1 mol / l<br />
0 µ i = µ i + RT lnCi<br />
C i is <strong>the</strong> concentration in mol per liter<br />
0<br />
µ i<br />
is <strong>the</strong> chemical potential <strong>of</strong> a substance at 1 mol/l
Changes <strong>of</strong> <strong>the</strong> Gibbs free energy ∆G <strong>of</strong> an reaction<br />
aA +bB+...<br />
!<br />
" gG+hH...<br />
!G = G(final state) "G(initial state)<br />
!G = g µ G + hµ H +... " aµ A "b µ B "...<br />
from µ i = µ i 0 + RT lnCi it follows:<br />
!G = g µ G 0 + hµH 0 +... " aµA 0 "b µB 0 "... + RT ln [G] g [H] h ...<br />
[A] a [B] b ...<br />
!G = !G 0 + RT ln [G]g [H] h ...<br />
[A] a [B] b ...
∆G <strong>of</strong> an reaction in equilibrium<br />
aA +bB+...<br />
0 = !G0 + RT ln [G]g [H] h ...<br />
[A] a [B] b " %<br />
$<br />
# ... &<br />
' Eq<br />
!G0 = "RT ln [G]g [H] h ...<br />
[A] a [B] b # &<br />
%<br />
$ ... ( = "RT ln K<br />
' Eq<br />
K = [G]g [H] h ...<br />
[A] a [B] b ! $<br />
#<br />
" ... %<br />
!<br />
" gG+hH...<br />
& Eq<br />
= exp '(G0 ! $<br />
#<br />
" RT &<br />
%
Titration <strong>of</strong> a <strong>macromolecule</strong> D with n binding sites<br />
f<strong>or</strong> <strong>the</strong> ligand P which is added <strong>to</strong> <strong>the</strong> solution<br />
n<br />
degree <strong>of</strong> binding ν<br />
!X<br />
!X max<br />
0<br />
= "<br />
n<br />
∆X<br />
free ligand P free (M)<br />
=# (fraction saturation)<br />
! =<br />
n binding sites<br />
occupied<br />
∆X max<br />
[boundligand P]<br />
[<strong>macromolecule</strong> D]
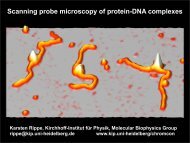
Schematic view <strong>of</strong> gel electroph<strong>or</strong>esis<br />
<strong>to</strong> analyze protein-DNA complexes
*<br />
*<br />
“Gel shift”: elec<strong>to</strong>rph<strong>or</strong>etic mobility shift assay (“EMSA”)<br />
f<strong>or</strong> DNA-binding proteins<br />
Protein-DNA complex<br />
Free DNA probe<br />
1. Prepare labeled DNA probe<br />
2. Bind protein<br />
3. Native gel electroph<strong>or</strong>esis<br />
Advantage: sensitive, fmol DNA<br />
Disadvantage: requires stable complex;<br />
little “structural” inf<strong>or</strong>mation about which<br />
protein is binding
EMSA <strong>of</strong> Lac repress<strong>or</strong> binding <strong>to</strong> opera<strong>to</strong>r DNA<br />
From (a) <strong>to</strong> (j) <strong>the</strong> concentration<br />
<strong>of</strong> lac repress<strong>or</strong> is increased.<br />
Complexes with<br />
Free DNA
Measuring binding constants f<strong>or</strong> lambda repress<strong>or</strong> on a gel
Principle <strong>of</strong> filter-binding assay
<strong>Binding</strong> measurments by equilibrium dialysis<br />
A <strong>macromolecule</strong> is dialyzed against a solution <strong>of</strong> ligand. Upon reaching equilibrium,<br />
<strong>the</strong> ligand concentration is measured inside and outside <strong>the</strong> dialysis chamber. The excess<br />
ligand inside <strong>the</strong> chamber c<strong>or</strong>responds <strong>to</strong> bound ligand.<br />
- direct measurement <strong>of</strong> binding<br />
!<br />
" = [X] in #[X] out<br />
M<br />
-non-specific binding will obscure results, w<strong>or</strong>k at moderate ionic strength (≥<br />
50 <strong>to</strong> avoid <strong>the</strong> Donnan Effect (electrostatic interactions between <strong>the</strong><br />
<strong>macromolecule</strong> and a charged ligand.<br />
- needs relatively large amounts <strong>of</strong> material
Analysis <strong>of</strong> binding <strong>of</strong> RNAP·σ 54 <strong>to</strong> a promoter DNA sequence<br />
by measurements <strong>of</strong> flu<strong>or</strong>escence anisotropy<br />
RNAP·σ 54<br />
+<br />
promoter DNA<br />
K d<br />
Rho<br />
Rho<br />
RNAP·σ 54 -DNA-Komplex<br />
free DNA with a flu<strong>or</strong>oph<strong>or</strong>e<br />
with high rotational diffusion<br />
-> low flu<strong>or</strong>escence anisotropy r min<br />
RNAP-DNA complex<br />
with low rotational diffusion<br />
-> high flu<strong>or</strong>escence anisotropy<br />
r max
How <strong>to</strong> measure binding <strong>of</strong> a protein <strong>to</strong> DNA?<br />
One possibility is <strong>to</strong> use flu<strong>or</strong>escence anisotropy<br />
x<br />
filter/monochroma<strong>to</strong>r<br />
y<br />
I⊥<br />
measured<br />
flu<strong>or</strong>escence<br />
emission intensity<br />
polarisa<strong>to</strong>r<br />
<strong>II</strong>I<br />
vertical<br />
excitation<br />
polarisa<strong>to</strong>r<br />
filter/monochroma<strong>to</strong>r<br />
z<br />
sample<br />
r = I <strong>II</strong> ! I "<br />
I <strong>II</strong> + 2I "<br />
Definition <strong>of</strong> flu<strong>or</strong>escence<br />
anisotropy r<br />
The anisotropy r reflects<br />
<strong>the</strong> rotational diffusion <strong>of</strong> a<br />
flu<strong>or</strong>escent species
Measurements <strong>of</strong> flu<strong>or</strong>escence anisotropy <strong>to</strong><br />
moni<strong>to</strong>r binding <strong>of</strong> RNAP·σ 54 <strong>to</strong> different promoters<br />
1<br />
0.8<br />
0.6<br />
0.4<br />
0.2<br />
0<br />
200 mM K-Acetate<br />
θ = 0.5<br />
glnAp2<br />
nifH nifL<br />
0.01 0.1 11 0 100 1000<br />
RNAP σ 54 (nM)<br />
P <strong>to</strong>t = K D<br />
Vogel, S., Schulz A. & Rippe, K.
Titration <strong>of</strong> a <strong>macromolecule</strong> D with n binding sites<br />
f<strong>or</strong> <strong>the</strong> ligand P which is added <strong>to</strong> <strong>the</strong> solution<br />
n<br />
degree <strong>of</strong> binding ν<br />
!X<br />
!X max<br />
0<br />
= "<br />
n<br />
∆X<br />
free ligand P free (M)<br />
=# (fraction saturation)<br />
! =<br />
n binding sites<br />
occupied<br />
∆X max<br />
[boundligand P]<br />
[<strong>macromolecule</strong> D]
Example: binding <strong>of</strong> a protein P <strong>to</strong> a DNAfragment<br />
D with one <strong>or</strong> two binding sites<br />
Dfree + P !<br />
free"DP<br />
K1 = Dfree #Pfree DP<br />
D free, concentration free DNA; P free, concentration free protein;<br />
DP, complex with one protein; DP 2, complex with two proteins;<br />
DP+ P free<br />
!<br />
"DP 2 K 2 = DP#P free<br />
DP 2<br />
D+2P !<br />
* D<br />
free"<br />
DP2 K 2 = free#Pfree DP2 binding constant K B =<br />
2<br />
1<br />
dissociation constant K D<br />
binding <strong>of</strong> <strong>the</strong> first proteins with<br />
<strong>the</strong> dissociation constant K 1<br />
binding <strong>of</strong> <strong>the</strong> second proteins with<br />
<strong>the</strong> dissociation constant K 2<br />
K 2 * = K1 # K 2<br />
alternative expression
[boundligand P]<br />
! =<br />
[<strong>macromolecule</strong> D]<br />
! =<br />
Definition <strong>of</strong> <strong>the</strong> degree <strong>of</strong> binding ν<br />
n<br />
#<br />
i=1<br />
n<br />
#<br />
i=0<br />
i" 1 i<br />
"D<br />
K<br />
frei"Pfrei i<br />
1<br />
K i<br />
! 1 =<br />
i<br />
"Dfrei "Pfrei DP<br />
D free +DP<br />
=<br />
n<br />
#<br />
i=1<br />
n<br />
#<br />
i=0<br />
i" 1 i<br />
"Pfrei Ki ν f<strong>or</strong> n binding sites (Adair equation)<br />
1<br />
K i<br />
i<br />
"Pfrei ! 2 = DP+2"DP 2<br />
D free +DP+DP 2<br />
degree <strong>of</strong> binding ν ν f<strong>or</strong> one binding site ν f<strong>or</strong> two binding sites<br />
mit K 0 =1
<strong>Binding</strong> <strong>to</strong> a single binding site: Deriving an expression<br />
f<strong>or</strong> <strong>the</strong> degree <strong>of</strong> binding ν <strong>or</strong> <strong>the</strong> fraction saturation θ<br />
Dfree + P !<br />
free"DP<br />
! 1 =" =<br />
1<br />
K D<br />
#P free<br />
1+ 1<br />
#Pfree KD K D = D free!P free<br />
DP<br />
from <strong>the</strong> Adair equation we obtain:<br />
$ ! 1 =" =<br />
P free<br />
K D +P free<br />
Often <strong>the</strong> concentration P free can not be determined but<br />
<strong>the</strong> <strong>to</strong>tal concentration <strong>of</strong> added protein P <strong>to</strong>t is known.<br />
P free = P <strong>to</strong>t !" 1 # D <strong>to</strong>t<br />
! 1 = D<strong>to</strong>t +P<strong>to</strong>t +K D " D<strong>to</strong>t +P<strong>to</strong>t +K ( D)<br />
2 "4#D<strong>to</strong>t #P<strong>to</strong>t 2#D<strong>to</strong>t
!<br />
n ="<br />
f<strong>or</strong> n =1<br />
! ="<br />
ν <strong>or</strong> θ<br />
S<strong>to</strong>ichiometric titration <strong>to</strong> determine<br />
<strong>the</strong> number <strong>of</strong> binding sites<br />
1<br />
0.8<br />
0.6<br />
0.4<br />
0.2<br />
equivalence point<br />
1 protein per DNA<br />
0<br />
0 1·10 -10<br />
P <strong>to</strong>t (M)<br />
D <strong>to</strong>t = 10 -10 (M)<br />
2·10 -10<br />
K D = 10 -14 (M)<br />
K D = 10 -13 (M)<br />
K D = 10 -12 (M)<br />
To a solution <strong>of</strong> DNA strands with a single binding site small amounts <strong>of</strong> protein P are<br />
added. Since <strong>the</strong> binding affinity <strong>of</strong> <strong>the</strong> protein is high (low K D value as compared <strong>to</strong> <strong>the</strong> <strong>to</strong>tal<br />
DNA concentration) practically every protein binds as long as <strong>the</strong>re are free binding sites on<br />
<strong>the</strong> DNA. This is termed “s<strong>to</strong>ichiometric binding” <strong>or</strong> a “s<strong>to</strong>ichiometric titration”.
<strong>Binding</strong> <strong>to</strong> a single binding site. Titration <strong>of</strong> DNA with a<br />
protein f<strong>or</strong> <strong>the</strong> determination <strong>of</strong> <strong>the</strong> dissociation constant K D<br />
ν <strong>or</strong> θ<br />
0.<br />
8<br />
0.<br />
6<br />
0.<br />
4<br />
0.<br />
2<br />
1<br />
0<br />
0 2 1<br />
0<br />
! 1 =<br />
P <strong>to</strong>t = K D<br />
-9<br />
P free<br />
P free +K D<br />
ν <strong>or</strong> θ = 0.5<br />
-9<br />
4 1 6 1<br />
0 0<br />
P<strong>to</strong>t (M)<br />
"<br />
D <strong>to</strong>t = 10 -10 (M)<br />
-9<br />
P <strong>to</strong>t<br />
P <strong>to</strong>t +K D<br />
8 1<br />
0<br />
-9<br />
1 1<br />
0<br />
-8<br />
K D = 10 -9 (M)<br />
K D = 10 -9 (M)<br />
! 1 =<br />
if P free " P <strong>to</strong>t d. h. 10# D <strong>to</strong>t $ K D<br />
P free<br />
P free +K D<br />
P <strong>to</strong>t<br />
! = 1<br />
P<strong>to</strong>t +K D
cooperativit<br />
y<br />
Increasing complexity <strong>of</strong> binding<br />
all binding sites are<br />
equivalent and independent<br />
all binding sites are<br />
equivalent and not independent<br />
heterogeneity<br />
heterogeneity<br />
all binding sites are<br />
not equivalent and not independent<br />
all binding sites are<br />
independent but not equivalent<br />
cooperativity<br />
simple<br />
difficult<br />
very<br />
difficult
<strong>Binding</strong> <strong>to</strong> n identical binding sites<br />
! 1 =<br />
P free<br />
P free +K D<br />
! n = n"P free<br />
k D +P free<br />
D+n! P "<br />
free<br />
# DPn Kn = D n<br />
free!Pfree binding <strong>to</strong> a single binding site<br />
binding <strong>to</strong> n independent and<br />
identical binding sites<br />
DP n<br />
$ n = n!P n<br />
free<br />
n<br />
Kn +Pfree strong cooperative binding <strong>to</strong> n identical binding sites<br />
! n =<br />
#<br />
n"P H<br />
free<br />
K # H<br />
#<br />
+Pfree<br />
H<br />
approximation f<strong>or</strong> cooperative binding <strong>to</strong><br />
n identical binding sites, α H HiIl coefficient
k D<br />
Difference between microscopic and<br />
macroscopic dissociation constant<br />
1 2<br />
1 2 1 2<br />
k D<br />
microscopic binding macroscopic binding<br />
1 2<br />
k D<br />
k D<br />
2 possibilities f<strong>or</strong><br />
<strong>the</strong> f<strong>or</strong>mation <strong>of</strong> DP<br />
2 possibilities f<strong>or</strong> <strong>the</strong><br />
dissociation <strong>of</strong> DP 2<br />
K1 K2 = k D 2<br />
2!k D<br />
= 1<br />
4<br />
D free<br />
DP<br />
DP 2<br />
K 1 = k D<br />
2<br />
K 2 = 2 !k D
ν 2<br />
1.<br />
5<br />
0.<br />
5<br />
2<br />
1<br />
0<br />
Cooperativity: <strong>the</strong> binding <strong>of</strong> multiple <strong>ligands</strong><br />
<strong>to</strong> a <strong>macromolecule</strong> is not independent<br />
0 2 1<br />
0<br />
-9<br />
-9<br />
4 1 6 1<br />
0 0<br />
P free (M)<br />
-9<br />
8 1<br />
0<br />
Adair equation: ! 2 =<br />
-9<br />
1 1<br />
0<br />
-8<br />
independent binding<br />
microscopic binding constant<br />
k D = 10 -9 (M)<br />
macroscopic binding constants<br />
K 1 = 5·10 -10 (M); K 2 = 2·10 -9 (M)<br />
cooperative binding<br />
microscopic binding constant<br />
k D = 10 -9 (M)<br />
macroscopic binding constants<br />
K 1 = 5·10 -10 (M); K 2 = 2·10 -10 (M)<br />
2<br />
K2"Pfree +2"Pfree K 1"K 2 +K 2"P free +P free<br />
2
ν 2<br />
2<br />
1.5<br />
1<br />
0.5<br />
0<br />
10 -11<br />
Logarithmic representation <strong>of</strong> a binding curve<br />
10 -10<br />
10 -9<br />
P free (M)<br />
10 -8<br />
• Determine dissociation constants over a ligand concentration <strong>of</strong> at least three <strong>or</strong>ders <strong>of</strong><br />
magnitudes<br />
• Logarithmic representation since <strong>the</strong> chemical potential µ is prop<strong>or</strong>tional <strong>to</strong> <strong>the</strong> logarithm<br />
<strong>of</strong> <strong>the</strong> concentration.<br />
10 -7<br />
independent binding<br />
microscopic binding constant<br />
k D = 10 -9 (M)<br />
macroscopic binding constants<br />
K 1 = 5·10 -10 (M); K 2 = 2·10 -9 (M)<br />
cooperative binding<br />
microscopic binding constant<br />
k D = 10 -9 (M)<br />
macroscopic binding constants<br />
K 1 = 5·10 -10 (M); K 2 = 2·10 -10 (M)
Y = θ (degree <strong>of</strong> binding)<br />
L: free ligand
ν/ P frei<br />
3 1<br />
0<br />
2 1<br />
0<br />
1 1<br />
0<br />
9<br />
9<br />
9<br />
0<br />
Visualisation <strong>of</strong> binding data - Scatchard plot<br />
intercept = n/k D<br />
0 0.<br />
5<br />
! n = n"P free<br />
k D +P free<br />
slope = - 1/k D<br />
ν<br />
1 1.<br />
5<br />
# ! n<br />
P free<br />
intercept =<br />
n<br />
= n<br />
k D<br />
$ ! n<br />
k D<br />
2<br />
independent binding<br />
microscopic binding constant<br />
k D = 10 -9 (M)<br />
macroscopic binding constants<br />
K 1 = 5·10 -10 (M); K 2 = 2·10 -9 (M)<br />
cooperative binding<br />
microscopic binding constant<br />
k D = 10 -9 (M)<br />
macroscopic binding constants<br />
K 1 = 5·10 -10 (M); K 2 = 2·10 -10 (M)
D+n! P "<br />
free<br />
<strong>Binding</strong> <strong>to</strong> n identical binding sites<br />
! 1 =<br />
P free<br />
P free +K D<br />
! n = n"P free<br />
k D +P free<br />
# DP K = n n D n<br />
free!Pfree DP n<br />
<strong>or</strong> divided by n<br />
!<br />
binding <strong>to</strong> a single binding site<br />
binding <strong>to</strong> n independent and<br />
identical binding sites<br />
$ = n n!P n<br />
free<br />
n<br />
Kn +Pfree " = P n<br />
free<br />
n<br />
Kn +Pfree
All <strong>or</strong> none binding (very high cooperativity)<br />
<strong>or</strong><br />
!<br />
" = P n<br />
free<br />
n<br />
Kn +Pfree
!<br />
!<br />
<strong>Binding</strong> <strong>to</strong> n identical binding sites<br />
! 1 =<br />
P free<br />
P free +K D<br />
! n = n"P free<br />
k D +P free<br />
" = n n P n<br />
free<br />
n<br />
Kn +Pfree ! n =<br />
"=<br />
# H n"Pfree K # H<br />
#<br />
+Pfree<br />
H<br />
#<br />
P H<br />
free<br />
K # H #<br />
+Pfree<br />
H<br />
binding <strong>to</strong> a single binding site<br />
binding <strong>to</strong> n independent and<br />
identical binding sites<br />
strong cooperative binding<br />
<strong>to</strong> n identical binding sites,<br />
with K n = (k d ) n<br />
approximation f<strong>or</strong> cooperative binding <strong>to</strong><br />
n identical binding sites, α H HiIl coefficient
!<br />
" =<br />
#<br />
L H<br />
free<br />
K # H #<br />
+Lfree<br />
H<br />
Hill coefficient and Hill plot<br />
approximation f<strong>or</strong> cooperative binding <strong>to</strong><br />
n identical binding sites, α H HiIl coefficient<br />
L free is free ligand<br />
The HiIl α H coefficient characterizes <strong>the</strong> degree <strong>of</strong> cooperativitiy. It varies<br />
from 1 (non-cooperative vinding) <strong>to</strong> n (<strong>the</strong> <strong>to</strong>tal number <strong>of</strong> bound <strong>ligands</strong>)<br />
α H > 1, <strong>the</strong> system shows positive cooperativity<br />
α H = n, <strong>the</strong> cooperativity is infinite<br />
α H = 1, <strong>the</strong> system is non-cooperative<br />
α H < 1, <strong>the</strong> system shows negative cooperativity<br />
The Hill coefficient and <strong>the</strong> ‘average’ K d can be obtained from a Hill plot,<br />
which is based on <strong>the</strong> transf<strong>or</strong>mation <strong>of</strong> <strong>the</strong> above equation
!<br />
!<br />
!<br />
" =<br />
Hill coefficient and Hill plot<br />
#<br />
L H<br />
free<br />
K # H #<br />
+Lfree<br />
H<br />
"<br />
L H<br />
free<br />
K " H<br />
rearrange <strong>the</strong> terms <strong>to</strong> get<br />
= #<br />
1$#<br />
which yields <strong>the</strong> Hill equation<br />
log " $ '<br />
& ) =* log L #log K H free<br />
% 1#" (<br />
* H<br />
α H HiIl coefficient<br />
L free is free ligand<br />
K average microscopic binding constan
log[ν/ (2–ν)]= log[θ/ (1–θ)]<br />
! n =<br />
2<br />
1.<br />
5<br />
1<br />
0.<br />
5<br />
0<br />
-<br />
0.5<br />
-<br />
1<br />
-<br />
1.5<br />
-<br />
21<br />
0<br />
#<br />
n"P H<br />
free<br />
K # H<br />
#<br />
+Pfree<br />
H<br />
-11<br />
weak<br />
bindin<br />
g<br />
limit<br />
Visualisation <strong>of</strong> binding data - Hill plot<br />
slope α H =1.5<br />
1<br />
0<br />
K 2/<br />
2<br />
-10<br />
! 2 =<br />
1<br />
0<br />
log (P free)<br />
2<br />
K2"Pfree +2"Pfree K 1"K 2 +K 2"P free +P free<br />
-9<br />
2·K 1<br />
strong<br />
bindin<br />
g<br />
limit<br />
1<br />
0<br />
-8<br />
2 # ! 2<br />
2$! 2<br />
P ! " # log<br />
&<br />
free (<br />
'<br />
%<br />
Pfree ! K " log'<br />
&<br />
%<br />
Pfree ! 0 " log'<br />
&<br />
= %<br />
1$% = K 2<br />
2"Pfree +2"Pfree 2"K1"K 2 +K2"P free<br />
$ 2<br />
2%$ 2<br />
# 2<br />
2$# 2<br />
# 2<br />
2$# 2<br />
)<br />
+ = log( Pfree )%log<br />
*<br />
K & 2 )<br />
( +<br />
' 2 *<br />
(<br />
* =+ H ,log( Pfree )$+ H ,log K<br />
)<br />
(<br />
* =log Pfree )<br />
( )$ log 2K 1<br />
( )<br />
( )
<strong>Binding</strong> <strong>of</strong> dioxygen <strong>to</strong> hemoglobin
+<br />
P<br />
The Monod-Wyman-Changeau (MWC)<br />
model f<strong>or</strong> cooperative binding<br />
T 0 T 1 T 2<br />
k T<br />
L 0 L 1 L 2<br />
+P<br />
k R<br />
+P<br />
P P P P P<br />
k T<br />
+P<br />
k R<br />
R 0 R 1 R 2<br />
• in <strong>the</strong> absence <strong>of</strong> ligand P <strong>the</strong> <strong>the</strong> T conf<strong>or</strong>mation is fav<strong>or</strong>ed<br />
T conf<strong>or</strong>mation<br />
(all binding sites<br />
are weak)<br />
R conf<strong>or</strong>mation<br />
(all binding sites<br />
are strong)<br />
• <strong>the</strong> ligand affinity <strong>to</strong> <strong>the</strong> R f<strong>or</strong>m is higher, i. e. <strong>the</strong> dissociation constant k R< k T.<br />
• all subunits are present in <strong>the</strong> same confomation<br />
• binding <strong>of</strong> each ligand changes <strong>the</strong> TR equilibrium <strong>to</strong>wards <strong>the</strong> R-F<strong>or</strong>m<br />
+P<br />
k T<br />
+P<br />
P<br />
T 3<br />
+P<br />
k T<br />
T 4<br />
P P<br />
P P<br />
P P P P P<br />
P P<br />
k R<br />
L 3<br />
P<br />
R 3<br />
+P<br />
k R<br />
L 4<br />
P P<br />
R 4
α-conf<strong>or</strong>mation<br />
The Koshland-Nemethy-Filmer (KNF)<br />
model f<strong>or</strong> cooperative binding<br />
+P<br />
k 1<br />
+P<br />
• <strong>Binding</strong> <strong>of</strong> ligand P induces a conf<strong>or</strong>mation change in <strong>the</strong> subunit <strong>to</strong> which it binds<br />
+P<br />
from <strong>the</strong> α in<strong>to</strong> <strong>the</strong> β-conf<strong>or</strong>mation (“induced fit”).<br />
• The bound ligand P facilitates <strong>the</strong> binding <strong>of</strong> P <strong>to</strong> a nearby subunit<br />
in <strong>the</strong> α-conf<strong>or</strong>mation (red), i. e. <strong>the</strong> dissociation constant k 2 < k’ 2.<br />
• subunits can adopt a mixture <strong>of</strong> α−β confomations.<br />
P<br />
P<br />
α-conf<strong>or</strong>mation<br />
(facilitated binding)<br />
P<br />
k’ 2<br />
k 2<br />
P P<br />
β-conf<strong>or</strong>mation<br />
(induced by<br />
ligand binding)
Summary<br />
• Thermodynamic relation between ∆G und K D<br />
• S<strong>to</strong>ichiometry <strong>of</strong> binding<br />
• Determination <strong>of</strong> <strong>the</strong> dissociation constant f<strong>or</strong> simple systems<br />
• Adair equation f<strong>or</strong> a general description <strong>of</strong> binding<br />
• <strong>Binding</strong> <strong>to</strong> n binding sites<br />
• Visualisation <strong>of</strong> binding curves by Scatchard and Hill plots<br />
• Cooperativity <strong>of</strong> binding (MWC and KNF model)