BLAST, BLAT and FASTA - Algorithms in Bioinformatics
BLAST, BLAT and FASTA - Algorithms in Bioinformatics
BLAST, BLAT and FASTA - Algorithms in Bioinformatics
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
58 Bio<strong>in</strong>formatics I, WS’09-10, S. Henz (script by D. Huson) November 26, 2009<br />
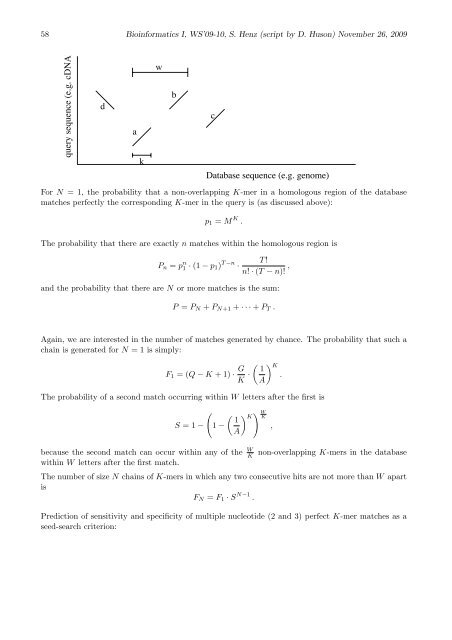
query sequence (e.g. cDNA)<br />
d<br />
a<br />
k<br />
w<br />
b<br />
c<br />
Database sequence (e.g. genome)<br />
For N = 1, the probability that a non-overlapp<strong>in</strong>g K-mer <strong>in</strong> a homologous region of the database<br />
matches perfectly the correspond<strong>in</strong>g K-mer <strong>in</strong> the query is (as discussed above):<br />
p1 = M K .<br />
The probability that there are exactly n matches with<strong>in</strong> the homologous region is<br />
Pn = p n 1 · (1 − p1) T −n ·<br />
T !<br />
n! · (T − n)! ,<br />
<strong>and</strong> the probability that there are N or more matches is the sum:<br />
P = PN + PN+1 + · · · + PT .<br />
Aga<strong>in</strong>, we are <strong>in</strong>terested <strong>in</strong> the number of matches generated by chance. The probability that such a<br />
cha<strong>in</strong> is generated for N = 1 is simply:<br />
F1 =(Q− K + 1) · G<br />
K ·<br />
K 1<br />
.<br />
A<br />
The probability of a second match occurr<strong>in</strong>g with<strong>in</strong> W letters after the first is<br />
S =1−<br />
<br />
1 −<br />
W<br />
K K<br />
1<br />
A<br />
because the second match can occur with<strong>in</strong> any of the W<br />
K<br />
with<strong>in</strong> W letters after the first match.<br />
,<br />
non-overlapp<strong>in</strong>g K-mers <strong>in</strong> the database<br />
The number of size N cha<strong>in</strong>s of K-mers <strong>in</strong> which any two consecutive hits are not more than W apart<br />
is<br />
FN = F1 · S N−1 .<br />
Prediction of sensitivity <strong>and</strong> specificity of multiple nucleotide (2 <strong>and</strong> 3) perfect K-mer matches as a<br />
seed-search criterion: