BLAST, BLAT and FASTA - Algorithms in Bioinformatics
BLAST, BLAT and FASTA - Algorithms in Bioinformatics
BLAST, BLAT and FASTA - Algorithms in Bioinformatics
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
54 Bio<strong>in</strong>formatics I, WS’09-10, S. Henz (script by D. Huson) November 26, 2009<br />
1. how many homologous regions are missed (FN), <strong>and</strong><br />
2. how many non-homologous regions are passed to the extension stage (FP), us<strong>in</strong>g this criteria,<br />
thus <strong>in</strong>creas<strong>in</strong>g the runn<strong>in</strong>g time of the application.<br />
4.11.2 Some def<strong>in</strong>itions<br />
K: The K-mer size<br />
M: Match ratio between homologous areas, ≈ 98% for cDNA/genomic alignments<br />
with<strong>in</strong> the same species, ≈ 89% for prote<strong>in</strong> alignments between human <strong>and</strong> mouse.<br />
H: The size of a homologous area. For a human exon this is typically 50 − 200 bp.<br />
G: Database size, e.g. 3 Gb for human.<br />
Q: Query size.<br />
A: Alphabet size, 20 for am<strong>in</strong>o acids, 4 for nucleotides.<br />
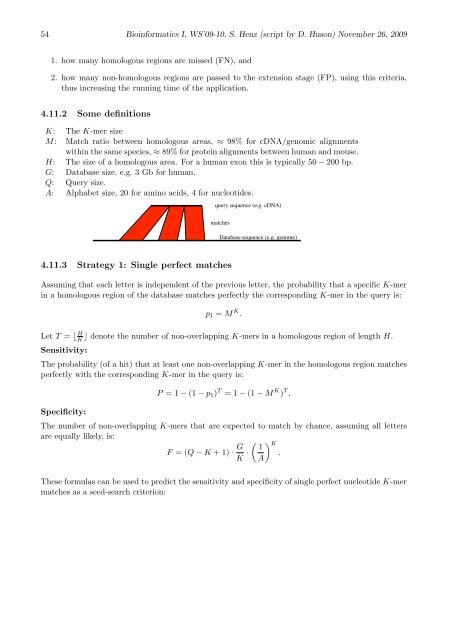
query sequence (e.g. cDNA)<br />
matches<br />
4.11.3 Strategy 1: S<strong>in</strong>gle perfect matches<br />
Database sequence (e.g. genome)<br />
Assum<strong>in</strong>g that each letter is <strong>in</strong>dependent of the previous letter, the probability that a specific K-mer<br />
<strong>in</strong> a homologous region of the database matches perfectly the correspond<strong>in</strong>g K-mer <strong>in</strong> the query is:<br />
p1 = M K .<br />
Let T = ⌊ H<br />
K ⌋ denote the number of non-overlapp<strong>in</strong>g K-mers <strong>in</strong> a homologous region of length H.<br />
Sensitivity:<br />
The probability (of a hit) that at least one non-overlapp<strong>in</strong>g K-mer <strong>in</strong> the homologous region matches<br />
perfectly with the correspond<strong>in</strong>g K-mer <strong>in</strong> the query is:<br />
Specificity:<br />
P =1− (1 − p1) T =1− (1 − M K ) T .<br />
The number of non-overlapp<strong>in</strong>g K-mers that are expected to match by chance, assum<strong>in</strong>g all letters<br />
are equally likely, is:<br />
F =(Q− K + 1) · G<br />
K ·<br />
K 1<br />
.<br />
A<br />
These formulas can be used to predict the sensitivity <strong>and</strong> specificity of s<strong>in</strong>gle perfect nucleotide K-mer<br />
matches as a seed-search criterion: