Free Radical Biomedicine: Principles, Clinical ... - Bentham Science
Free Radical Biomedicine: Principles, Clinical ... - Bentham Science
Free Radical Biomedicine: Principles, Clinical ... - Bentham Science
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
Detection of Antioxidants <strong>Free</strong> <strong>Radical</strong> <strong>Biomedicine</strong>: <strong>Principles</strong>, <strong>Clinical</strong> Correlations, and Methodologies 309<br />
common cellular antioxidants with an emphasis on the principles and major procedures of the assays for<br />
measuring antioxidant enzymes.<br />
2. MAJOR EXPERIMENTAL APPROACHES TO ANALYZING ANTIOXIDANTS<br />
As stated in Chapter 3, mammalian cells and tissues are equipped with a range of antioxidants, including<br />
both protein and non-protein antioxidants synthesized by cells. In addition, mammalian antioxidant<br />
defenses also comprise the antioxidant compounds (e.g., vitamin C, vitamin E, carotenoids, and<br />
polyphenols) derived from the diet. Although the proper functionality of mammalian cells depends on the<br />
coordinated actions of these diverse dietary antioxidants, endogenous antioxidant enzymes seem to play a<br />
more vital role in protecting against oxidative stress. This is evidenced by the essentiality of some<br />
antioxidant enzymes in embryonic development as well as the disease phenotypes caused by deletion of<br />
each of the many cellular antioxidant genes in animal models. Hence, methodologies have been developed<br />
to specifically detect and measure cellular and tissue antioxidant enzymes and proteins. Antioxidant<br />
enzymes and proteins can be assessed by a number of major experimental approaches, as summarized<br />
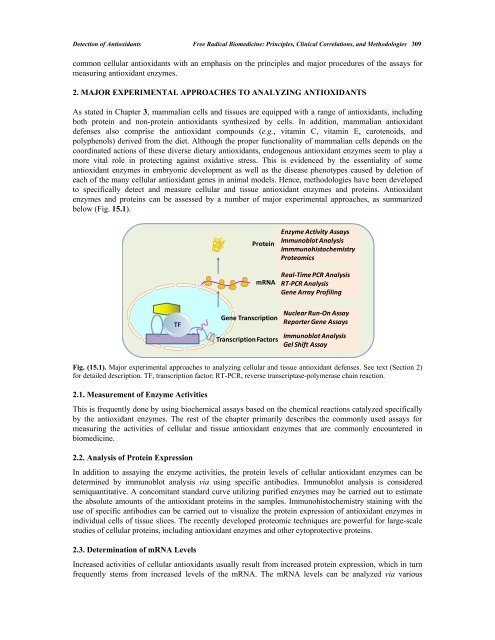
below (Fig. 15.1).<br />
TF<br />
Protein<br />
mRNA<br />
Gene Transcription<br />
Transcription Factors<br />
Enzyme Activity Assays<br />
Immunoblot Analysis<br />
Immmunohistochemistry<br />
Proteomics<br />
Real‐Time PCR Analysis<br />
RT‐PCR Analysis<br />
Gene Array Profiling<br />
Nuclear Run‐On Assay<br />
Reporter Gene Assays<br />
Immunoblot Analysis<br />
Gel Shift Assay<br />
Fig. (15.1). Major experimental approaches to analyzing cellular and tissue antioxidant defenses. See text (Section 2)<br />
for detailed description. TF, transcription factor; RT-PCR, reverse transcriptase-polymerase chain reaction.<br />
2.1. Measurement of Enzyme Activities<br />
This is frequently done by using biochemical assays based on the chemical reactions catalyzed specifically<br />
by the antioxidant enzymes. The rest of the chapter primarily describes the commonly used assays for<br />
measuring the activities of cellular and tissue antioxidant enzymes that are commonly encountered in<br />
biomedicine.<br />
2.2. Analysis of Protein Expression<br />
In addition to assaying the enzyme activities, the protein levels of cellular antioxidant enzymes can be<br />
determined by immunoblot analysis via using specific antibodies. Immunoblot analysis is considered<br />
semiquantitative. A concomitant standard curve utilizing purified enzymes may be carried out to estimate<br />
the absolute amounts of the antioxidant proteins in the samples. Immunohistochemistry staining with the<br />
use of specific antibodies can be carried out to visualize the protein expression of antioxidant enzymes in<br />
individual cells of tissue slices. The recently developed proteomic techniques are powerful for large-scale<br />
studies of cellular proteins, including antioxidant enzymes and other cytoprotective proteins.<br />
2.3. Determination of mRNA Levels<br />
Increased activities of cellular antioxidants usually result from increased protein expression, which in turn<br />
frequently stems from increased levels of the mRNA. The mRNA levels can be analyzed via various