The role of human and Drosophila NXF proteins in nuclear mRNA ...
The role of human and Drosophila NXF proteins in nuclear mRNA ...
The role of human and Drosophila NXF proteins in nuclear mRNA ...
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
Introduction 26<br />
experiments revealed that the presence <strong>of</strong> a poly(A) tail does not significantly enhance the<br />
export efficiency <strong>of</strong> these transcripts (see above; Jarmolowski et al., 1994).<br />
Another l<strong>in</strong>k between <strong>nuclear</strong> <strong>mRNA</strong> export <strong>and</strong> 3' end formation has been made<br />
by analyz<strong>in</strong>g transcripts <strong>in</strong> yeast stra<strong>in</strong>s defective <strong>in</strong> <strong>mRNA</strong> export. Defects <strong>in</strong> RAT7<br />
(Nup159), GLE1, MEX67, RAT8 (Dbp5) or RIP1 genes, which all result <strong>in</strong> a block to<br />
<strong>mRNA</strong> export, cause hyperadenylation <strong>of</strong> transcripts (Hammell et al., 2002; Hilleren <strong>and</strong><br />
Parker, 2001; Jensen et al., 2001b). Furthermore, 3'-extended <strong>mRNA</strong>s (result<strong>in</strong>g from<br />
improper transcription term<strong>in</strong>ation) are frequently observed <strong>in</strong> <strong>mRNA</strong> export mutant<br />
stra<strong>in</strong>s (Hammell et al., 2002). <strong>The</strong>se 3'-extended <strong>and</strong>/or hyperadenylated <strong>mRNA</strong>s<br />
accumulate at the site <strong>of</strong> transcription. Similarly, <strong>in</strong>completely processed transcripts also<br />
accumulate at the site <strong>of</strong> transcription <strong>in</strong> mammalian cells (Custodio et al., 1999; Hilleren<br />
et al., 2001; Jensen et al., 2001b; Libri et al., 2002). Interest<strong>in</strong>gly, not all yeast <strong>mRNA</strong><br />
export mutants show defects <strong>in</strong> 3' end formation: yra1 or sub2 mutants neither display<br />
term<strong>in</strong>ation nor polyadenylation defects (Hammell et al., 2002).<br />
Even though these data <strong>in</strong>dicate a coupl<strong>in</strong>g between 3' end formation <strong>and</strong> <strong>mRNA</strong><br />
export provid<strong>in</strong>g an efficient retention mechanism <strong>of</strong> <strong>in</strong>completely processed transcripts,<br />
the molecular basis underly<strong>in</strong>g this phenomenon rema<strong>in</strong>s to be established.<br />
Other factors implicated <strong>in</strong> <strong>mRNA</strong> export<br />
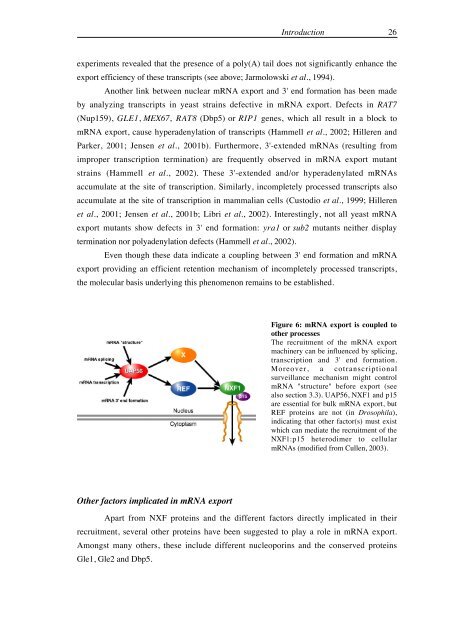
Figure 6: <strong>mRNA</strong> export is coupled to<br />
other processes<br />
<strong>The</strong> recruitment <strong>of</strong> the <strong>mRNA</strong> export<br />
mach<strong>in</strong>ery can be <strong>in</strong>fluenced by splic<strong>in</strong>g,<br />
transcription <strong>and</strong> 3' end formation.<br />
Moreover, a cotranscriptional<br />
surveillance mechanism might control<br />
<strong>mRNA</strong> "structure" before export (see<br />
also section 3.3). UAP56, <strong>NXF</strong>1 <strong>and</strong> p15<br />
are essential for bulk <strong>mRNA</strong> export, but<br />
REF <strong>prote<strong>in</strong>s</strong> are not (<strong>in</strong> <strong>Drosophila</strong>),<br />
<strong>in</strong>dicat<strong>in</strong>g that other factor(s) must exist<br />
which can mediate the recruitment <strong>of</strong> the<br />
<strong>NXF</strong>1:p15 heterodimer to cellular<br />
<strong>mRNA</strong>s (modified from Cullen, 2003).<br />
Apart from <strong>NXF</strong> <strong>prote<strong>in</strong>s</strong> <strong>and</strong> the different factors directly implicated <strong>in</strong> their<br />
recruitment, several other <strong>prote<strong>in</strong>s</strong> have been suggested to play a <strong>role</strong> <strong>in</strong> <strong>mRNA</strong> export.<br />
Amongst many others, these <strong>in</strong>clude different nucleopor<strong>in</strong>s <strong>and</strong> the conserved <strong>prote<strong>in</strong>s</strong><br />
Gle1, Gle2 <strong>and</strong> Dbp5.