What stabilizes the N.A. double helix? < *
What stabilizes the N.A. double helix? < *
What stabilizes the N.A. double helix? < *
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
*<br />
*<br />
*<br />
<strong>What</strong> <strong>stabilizes</strong> <strong>the</strong> N.A. <strong>double</strong> <strong>helix</strong>?<br />
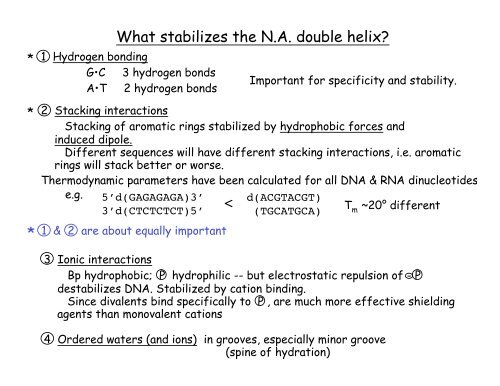
1 Hydrogen bonding<br />
G•C 3 hydrogen bonds<br />
A•T 2 hydrogen bonds<br />
2 Stacking interactions<br />
Stacking of aromatic rings stabilized by hydrophobic forces and<br />
induced dipole.<br />
Different sequences will have different stacking interactions, i.e. aromatic<br />
rings will stack better or worse.<br />
Thermodynamic parameters have been calculated for all DNA & RNA dinucleotides.<br />
e.g.<br />
5’d(GAGAGAGA)3’<br />
3’d(CTCTCTCT)5’<br />
1 & 2 are about equally important<br />
Important for specificity and stability.<br />
< d(ACGTACGT)<br />
(TGCATGCA)<br />
T m<br />
~20° different<br />
3 Ionic interactions<br />
Bp hydrophobic; P hydrophilic -- but electrostatic repulsion of - P<br />
de<strong>stabilizes</strong> DNA. Stabilized by cation binding.<br />
Since divalents bind specifically to P , are much more effective shielding<br />
agents than monovalent cations<br />
4 Ordered waters (and ions) in grooves, especially minor groove<br />
(spine of hydration)
DNA denaturation<br />
DNA bases absorb UV light due to aromatic rings.<br />
λ max<br />
at A 260<br />
(average for <strong>the</strong> four bases at pH 7.0)<br />
Recall: A = εcl<br />
path length, usually 1 cm<br />
1 mg/ml DNA has A ~<br />
260 = 20<br />
50 µg/ml DNA has A ~<br />
260 = 1<br />
absorbance<br />
extinction<br />
coefficient<br />
concentration<br />
Hyperchromic effect: stacking of nucleotide bases decreases ε.<br />
∴ bases in ss absorb more than bases in ds.<br />
⇒ Absorbance increases as DNA denatures.<br />
ds → ss, A 260 ↑ ~37%<br />
transition breadth<br />
T m<br />
(melting temperature) is transition midpoint<br />
[wc] = [w] = [c] half melted<br />
Approximate inflection point on melting curve
DNA melting is a cooperative process<br />
Increasing Conformational Entropy<br />
Increasing translational<br />
Entropy
<strong>What</strong> makes a process cooperative?<br />
First, it must occur in multiple steps. For example:<br />
K<br />
A<br />
1<br />
K<br />
B<br />
2<br />
K<br />
C<br />
3<br />
D<br />
Second, <strong>the</strong> equilibrium constants of <strong>the</strong> successive steps must<br />
satisfy <strong>the</strong> following relationship:<br />
K 1 < K 2 < K 3
Stability of DNA & ∴ T m<br />
are affected by:<br />
1) ionic strength of solution<br />
2) GC content of DNA<br />
3) pH<br />
4) solvent<br />
5) binding of molecules<br />
(e.g. drugs, proteins, poly cations)<br />
1) Ionic strength:<br />
µ = (1/2)c i<br />
z i<br />
2<br />
charge of each species<br />
concentration of each species<br />
for a given ion, as [cation] ↑, T m<br />
↑<br />
2) GC content:<br />
@ same ionic strength, DNAs with<br />
higher GC content will melt higher<br />
[Exception: short DNA oligonucleotides ≤ 50 bp]<br />
Combine 1) & 2) to get empirical equation for Na + :<br />
T m<br />
= 41.1 (X G+C<br />
) + 16.6 log[Na + ] + 81.5<br />
mole fraction<br />
(know how to use,<br />
but don’t need to memorize)
Relationship between GC content and T m<br />
Original Marmur/Doty equation:<br />
T m<br />
= 0.411 (%GC) + 69<br />
%GC = 100χ GC<br />
T m<br />
= 41.1 χ GC<br />
+ 69<br />
Marmur and Doty<br />
(1962) JMB 5, 119-131<br />
Experimental observation: 10-fold<br />
increase in [Na + ] increases T m<br />
by 16.6˚.<br />
This leads to <strong>the</strong> salt corrected<br />
Marmur/Doty equation:<br />
T m<br />
= 41.1 χ GC<br />
+ 16.6log [Na + ] + 81.5
Marmur/Doty relationship breaks down<br />
for small and/or non-complex duplexes<br />
Sequence GC Exp T m M/D T m<br />
GGATCC<br />
CCTAGG<br />
CAAGCTTG<br />
GTTCGAAC<br />
GGTATACC<br />
CCATATGG<br />
0.67 30.8 C 109 C<br />
0.50 44.6 C 103<br />
0.50 36.0 C 103<br />
C<br />
C<br />
Discrepancies with Marmur/Doty equation are due to length<br />
effects and nearest neighbor effects.
Renaturation of DNA<br />
Melted DNA can be reannealed<br />
to an extent which depends on<br />
sequence complexity & rate of reannealing.<br />
e.g.<br />
Rate: If cools too fast, strands don’t have time to find neighbors<br />
Sequence Complexity: For random DNA (e.g. calf thymus, historical source)<br />
don’t get much reannealing. For poly d(AT) n<br />
, can get complete reannealing<br />
Historically important for finding highly repetitive DNA sequences in chromatin,<br />
e.g. satellite DNA (before DNA sequencing).<br />
Important today in molecular biology experiments,<br />
e.g. DNA or RNA hybridization (Nor<strong>the</strong>rn blots, Sou<strong>the</strong>rn blots, PCR)