Book of abstracts - State Scientific Institution “Institute for Single ...
Book of abstracts - State Scientific Institution “Institute for Single ...
Book of abstracts - State Scientific Institution “Institute for Single ...
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
Abstracts: Lectures<br />
LECT-19<br />
Detection <strong>of</strong> unlabelled oligonucleotide targets using<br />
whispering gallery modes in single, fluorescent microspheres<br />
Edin Nuhiji, Paul Mulvaney<br />
School <strong>of</strong> Chemistry & Bio21 Institute Level 2 North, 30 Flemington Road, University <strong>of</strong> Melbourne<br />
Parkville, VIC, 3010 (Australia). E-mail: <br />
Sequence specific nucleic acid detection has become an important goal in an array <strong>of</strong> biotechnology and<br />
biomedical disciplines as well as in <strong>for</strong>ensic analysis. However a disadvantage <strong>of</strong> most current nucleic acid<br />
detection systems is the necessity to label the analyte. [1, 2] Labelling the target molecule can be costly,<br />
laborious and may interfere with the binding affinity <strong>of</strong> the target molecule. In this work we demonstrate<br />
the development and characterization <strong>of</strong> an innovative, highly-sensitive, label-free oligonucleotide specific<br />
biosensor. [3]<br />
Whispering gallery modes (WGM) have emerged as a powerful signal transduction mechanism that could<br />
be utilized in ultra-sensitive biosensors. [4] In dielectric microspheres WGM are produced when incident<br />
radiation is internally trapped as standing waves, resulting in a scattering spectrum composed <strong>of</strong> numerous,<br />
sharp peaks. Light is introduced into the microsphere by coupling through an optic fibre or more simply by<br />
attaching a fluorescent molecule to a microspheres surface and illuminating with a UV lamp. The<br />
fluorescent microspheres within this work comprise a silica microsphere functionalised with a fluorophore<br />
and a dense monolayer <strong>of</strong> 5’ tethered, single-stranded oligonucleotides. The adsorption <strong>of</strong> the unlabelled<br />
complementary (cDNA) probe then causes nanometre shifts in the emission spectrum <strong>of</strong> the microsphere.<br />
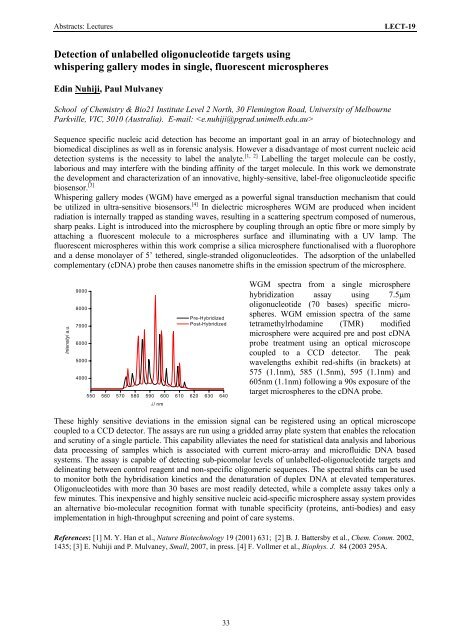
Intensity/ a.u.<br />
9000<br />
8000<br />
7000<br />
6000<br />
5000<br />
4000<br />
550 560 570 580 590 600 610 620 630 640<br />
λ/ nm<br />
Pre-Hybridized<br />
Post-Hybridized<br />
WGM spectra from a single microsphere<br />
hybridization assay using 7.5μm<br />
oligonucleotide (70 bases) specific microspheres.<br />
WGM emission spectra <strong>of</strong> the same<br />
tetramethylrhodamine (TMR) modified<br />
microsphere were acquired pre and post cDNA<br />
probe treatment using an optical microscope<br />
coupled to a CCD detector. The peak<br />
wavelengths exhibit red-shifts (in brackets) at<br />
575 (1.1nm), 585 (1.5nm), 595 (1.1nm) and<br />
605nm (1.1nm) following a 90s exposure <strong>of</strong> the<br />
target microspheres to the cDNA probe.<br />
These highly sensitive deviations in the emission signal can be registered using an optical microscope<br />
coupled to a CCD detector. The assays are run using a gridded array plate system that enables the relocation<br />
and scrutiny <strong>of</strong> a single particle. This capability alleviates the need <strong>for</strong> statistical data analysis and laborious<br />
data processing <strong>of</strong> samples which is associated with current micro-array and micr<strong>of</strong>luidic DNA based<br />
systems. The assay is capable <strong>of</strong> detecting sub-picomolar levels <strong>of</strong> unlabelled-oligonucleotide targets and<br />
delineating between control reagent and non-specific oligomeric sequences. The spectral shifts can be used<br />
to monitor both the hybridisation kinetics and the denaturation <strong>of</strong> duplex DNA at elevated temperatures.<br />
Oligonucleotides with more than 30 bases are most readily detected, while a complete assay takes only a<br />
few minutes. This inexpensive and highly sensitive nucleic acid-specific microsphere assay system provides<br />
an alternative bio-molecular recognition <strong>for</strong>mat with tunable specificity (proteins, anti-bodies) and easy<br />
implementation in high-throughput screening and point <strong>of</strong> care systems.<br />
References: [1] M. Y. Han et al., Nature Biotechnology 19 (2001) 631; [2] B. J. Battersby et al., Chem. Comm. 2002,<br />
1435; [3] E. Nuhiji and P. Mulvaney, Small, 2007, in press. [4] F. Vollmer et al., Biophys. J. 84 (2003 295A.<br />
33







![Changes in crystalli] ation conditions when growing large single ...](https://img.yumpu.com/23526254/1/184x260/changes-in-crystalli-ation-conditions-when-growing-large-single-.jpg?quality=85)






