X-ray Structures and Analysis of 11 Cyclosporin Derivatives ...
X-ray Structures and Analysis of 11 Cyclosporin Derivatives ...
X-ray Structures and Analysis of 11 Cyclosporin Derivatives ...
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
436 <strong>Cyclosporin</strong> <strong>Derivatives</strong> Complexed with CypA<br />
Table 1. Formulae <strong>of</strong> CsA analogues <strong>and</strong> their biological<br />
activities<br />
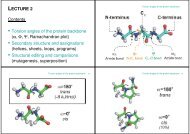
Figure 1. Residue labelling scheme for CsA-analogues<br />
(see also Table 1). Atom labelling is according to the<br />
IUPAC convention. CN is the methyl carbon atom <strong>of</strong><br />
the N-methylated amino acids. MLE for MeLeu; MVA<br />
for MeVal; BMT for MeBmt; ABU for Abu; SAR for Sar;<br />
VAL for Val; ALA for Ala; DAL for D-Ala. All amino<br />
acid residues with the exception <strong>of</strong> D-Ala8 (<strong>and</strong> Sar3)<br />
are in the L-con®guration.<br />
activity (Sigal et al., 1991). Even small chemical<br />
changes to residues <strong>of</strong> the effector loop can destroy<br />
the immunsuppressive effect without reducing the<br />
ability to bind cyclophilin (Papageorgiou et al.,<br />
1994a).<br />
This paper describes the structures <strong>of</strong> eight<br />
chemically distinct CsA-analogues complexed with<br />
CypA which show modi®cations in both the cyclophilin-binding<br />
residues <strong>and</strong> the effector loop.<br />
A comparison <strong>of</strong> each complex with the native<br />
CypA/CsA structure shows differences in conformation,<br />
molecular rigidity <strong>and</strong> water binding. This<br />
small library <strong>of</strong> closely related lig<strong>and</strong> structures<br />
also provides a picture <strong>of</strong> the frequently unpredictable<br />
effects <strong>of</strong> small chemical changes on 3D structure<br />
<strong>and</strong> biological activity.<br />
Results<br />
Side-chains for the amino acids at position 1, 2, 3 <strong>and</strong> 4 are<br />
shown for the <strong>11</strong> different CsA derivative complexes. The horizontal<br />
line represents the Ca-C b bond for the side-chain. The<br />
CypA value in column 6 gives a measure <strong>of</strong> the strength <strong>of</strong><br />
binding <strong>of</strong> the derivative relative to CsA value in column 6<br />
gives a measure <strong>of</strong> the strength <strong>of</strong> binding <strong>of</strong> the derivative<br />
relative to CsA as determined using an ELISA assay<br />
(Quesniaux et al., 1988). The measure <strong>of</strong> the effect <strong>of</strong> suppressing<br />
the production <strong>of</strong> interleukin-2 relative to CsA in a whole<br />
cell assay is given in the column labeled IL2 (Fliri et al., 1993;<br />
Bollinger et al., 1990).<br />
CsA ˆ Abu2-CS; <strong>11</strong>6450 ˆ MeBm 2 t-CS; 33804 ˆ Val2-CS;<br />
27402 ˆ Thr2-CS; 224698 ˆ (5-hydroxy)Nva2-CS; 209313 ˆ<br />
D-MeSer3-CS; 209650 ˆ Val2-D-MeAla3-CS; 209217 ˆ Val2-D-(2-<br />
S-methyl) Sar3-CS; 209825 ˆ (6,7-dihydro)MeBmt-1-Val2-D-(2-<br />
S-methyl)Sar3-CS; 2<strong>11</strong>810 ˆ (4-hydroxy)MeLeu4-CS; 2<strong>11</strong>8<strong>11</strong> ˆ<br />
MeIle4-CS.<br />
a IC 50 (derivative)/IC 50 CsA.<br />
b 26 times less binding than CsA.<br />
c 4.2 times less immunosuppressive than CsA.<br />
d Three times better binding than CsA.<br />
X-<strong>ray</strong> results<br />
X-<strong>ray</strong> structures <strong>of</strong> a total <strong>of</strong> <strong>11</strong> cyclosporines cocrystallised<br />
with cyclophilin are presented. The labelling<br />
scheme is shown in Figure 1 <strong>and</strong> chemical<br />
structures <strong>of</strong> the cyclosporines are described in<br />
Table 1. For completeness, these include three previously<br />
published structures: native CsA (Mikol<br />
et al., 1993), <strong>11</strong>6450 ((4-methyl)MeBmt1-CS) (Mikol<br />
et al., 1994) <strong>and</strong> 224698 ((5-hydroxy)Nva2-CS)<br />
(Mikol et al., 1995). Most <strong>of</strong> the CypA/CsA-analogue<br />
complexes were grown using a cross-seeding<br />
technique (Mikol & Duc, 1994). These crystals<br />
grew isomorphously with space group P2 1 2 1 2 1<br />
<strong>and</strong> cell dimensions a ˆ 36.4 AÊ , b ˆ 60.7 AÊ ,<br />
c ˆ 72.2 AÊ . The re®ned cell dimensions for the<br />
different isomorphous complexes containing CsAanalogues<br />
varied by a maximum <strong>of</strong> 0.6 AÊ in the a-<br />
dimension, 1.6 AÊ in the b-dimension <strong>and</strong> 1.4 AÊ in<br />
the c-dimension (see Table 2). The unlig<strong>and</strong>ed<br />
CypA structure showed a shrinkage <strong>of</strong> over 2 AÊ<br />
in the b <strong>and</strong> c-dimensions with a ˆ 36.46 AÊ ,<br />
b ˆ 57.32 AÊ , c ˆ 70.73 AÊ . The complex CypA/<br />
209313 was crystallised in space group P2 1 2 1 2<br />
with unit cell dimensions a ˆ 62.9 AÊ , b ˆ 65.3 AÊ ,<br />
c ˆ 40.8 AÊ .<br />
The <strong>11</strong> structures are re®ned to a resolution <strong>of</strong><br />
between 2.2 AÊ <strong>and</strong> 1.76 AÊ . <strong>and</strong> give R-factors