Simulations of Biomolecules Using Molecular Dynamics
Simulations of Biomolecules Using Molecular Dynamics
Simulations of Biomolecules Using Molecular Dynamics
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
2<br />
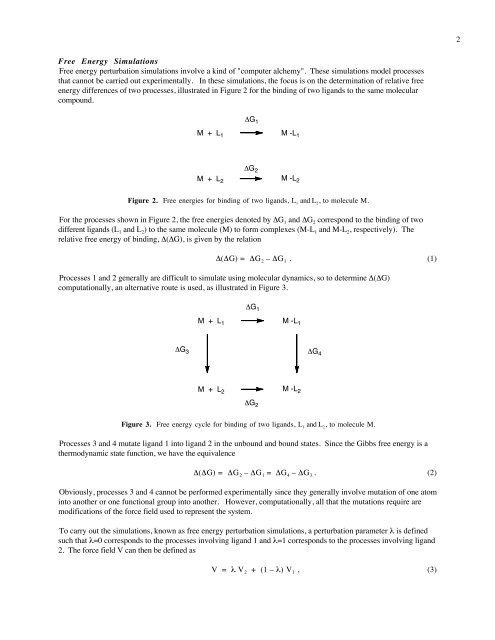
Free Energy <strong>Simulations</strong><br />
Free energy perturbation simulations involve a kind <strong>of</strong> "computer alchemy". These simulations model processes<br />
that cannot be carried out experimentally. In these simulations, the focus is on the determination <strong>of</strong> relative free<br />
energy differences <strong>of</strong> two processes, illustrated in Figure 2 for the binding <strong>of</strong> two ligands to the same molecular<br />
compound.<br />
ΔG 1<br />
M + L 1 M -L 1<br />
ΔG 2<br />
M + L 2<br />
M -L 2<br />
Figure 2. Free energies for binding <strong>of</strong> two ligands, L 1 and L 2 , to molecule M.<br />
For the processes shown in Figure 2, the free energies denoted by ΔG 1 and ΔG 2 correspond to the binding <strong>of</strong> two<br />
different ligands (L 1 and L 2 ) to the same molecule (M) to form complexes (M-L 1 and M-L 2 , respectively). The<br />
relative free energy <strong>of</strong> binding, Δ(ΔG), is given by the relation<br />
Δ(ΔG) = ΔG 2 – ΔG 1 . (1)<br />
Processes 1 and 2 generally are difficult to simulate using molecular dynamics, so to determine Δ(ΔG)<br />
computationally, an alternative route is used, as illustrated in Figure 3.<br />
ΔG 1<br />
M + L 1 M -L 1<br />
M + L 2<br />
M -L 2<br />
ΔG 2<br />
Figure 3. Free energy cycle for binding <strong>of</strong> two ligands, L 1 and L 2 , to molecule M.<br />
Processes 3 and 4 mutate ligand 1 into ligand 2 in the unbound and bound states. Since the Gibbs free energy is a<br />
thermodynamic state function, we have the equivalence<br />
Δ(ΔG) = ΔG 2 – ΔG 1 = ΔG 4 – ΔG 3 . (2)<br />
Obviously, processes 3 and 4 cannot be performed experimentally since they generally involve mutation <strong>of</strong> one atom<br />
into another or one functional group into another. However, computationally, all that the mutations require are<br />
modifications <strong>of</strong> the force field used to represent the system.<br />
To carry out the simulations, known as free energy perturbation simulations, a perturbation parameter λ is defined<br />
such that λ=0 corresponds to the processes involving ligand 1 and λ=1 corresponds to the processes involving ligand<br />
2. The force field V can then be defined as<br />
V = λ V 2 + (1 – λ) V 1 , (3)