Fuzzy c-means
Fuzzy c-means
Fuzzy c-means
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
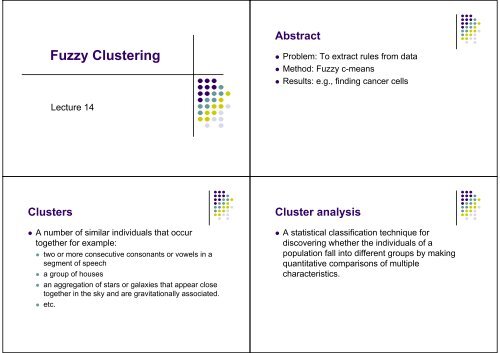
Abstract<br />
<strong>Fuzzy</strong> Clustering<br />
• Problem: To extract rules from data<br />
• Method: <strong>Fuzzy</strong> c-<strong>means</strong><br />
• Results: e.g., finding cancer cells<br />
Lecture 14<br />
Clusters<br />
• A number of similar individuals that occur<br />
together for example:<br />
• two or more consecutive consonants or vowels in a<br />
segment of speech<br />
• a group of houses<br />
• an aggregation of stars or galaxies that appear close<br />
together in the sky and are gravitationally associated.<br />
• etc.<br />
Cluster analysis<br />
• A statistical classification technique for<br />
discovering whether the individuals of a<br />
population fall into different groups by making<br />
quantitative comparisons of multiple<br />
characteristics.
Vehicle Example<br />
Vehicle Clusters<br />
Vehicle Top speed<br />
km/h<br />
Colour Air<br />
resistance<br />
Weight<br />
Kg<br />
V1 220 red 0.30 1300<br />
V2 230 black 0.32 1400<br />
V3 260 red 0.29 1500<br />
V4 140 gray 0.35 800<br />
V5 155 blue 0.33 950<br />
V6 130 white 0.40 600<br />
V7 100 black 0.50 3000<br />
V8 105 red 0.60 2500<br />
V9 110 gray 0.55 3500<br />
Weight [kg]<br />
3500<br />
3000<br />
2500<br />
2000<br />
1500<br />
1000<br />
Lorries<br />
Medium market cars<br />
Sports cars<br />
500<br />
100 150 200 250 300<br />
Top speed [km/h]<br />
Terminology<br />
feature<br />
Weight [kg]<br />
3500<br />
3000<br />
2500<br />
2000<br />
1500<br />
1000<br />
Object or data point<br />
Lorries<br />
Medium market cars<br />
Sports cars<br />
500<br />
100 150 200 250 300<br />
Top speed [km/h]<br />
feature<br />
cluster<br />
label<br />
feature space<br />
Classify cracked tiles<br />
475Hz 557Hz Ok<br />
-----+-----+---<br />
0.958 0.003 Yes<br />
1.043 0.001 Yes<br />
1.907 0.003 Yes<br />
0.780 0.002 Yes<br />
0.579 0.001 Yes<br />
0.003 0.105 No<br />
0.001 1.748 No<br />
0.014 1.839 No<br />
0.007 1.021 No<br />
0.004 0.214 No<br />
Table 1: frequency<br />
intensities for ten<br />
tiles.<br />
Tiles are made from clay moulded into the right shape, brushed, glazed, and baked.<br />
Unfortunately, the baking may produce invisible cracks. Operators can detect the<br />
cracks by hitting the tiles with a hammer, and in an automated system the response is<br />
recorded with a microphone, filtered, Fourier transformed, and normalised. A small set<br />
of data is given in TABLE 1 (adapted from MIT, 1997).
hard c-<strong>means</strong> (HCM)<br />
(also known as k <strong>means</strong>)<br />
2<br />
Tiles data: o = whole tiles, * = cracked tiles, x = centres<br />
2<br />
Tiles data: o = whole tiles, * = cracked tiles, x = centres<br />
1<br />
1<br />
0<br />
0<br />
log(intensity) 557 Hz<br />
-1<br />
-2<br />
-3<br />
-4<br />
-5<br />
log(intensity) 557 Hz<br />
-1<br />
-2<br />
-3<br />
-4<br />
-5<br />
-6<br />
-6<br />
-7<br />
-7<br />
-8<br />
-8 -6 -4 -2 0 2<br />
log(intensity) 475 Hz<br />
Plot of tiles by frequencies (logarithms). The whole tiles (o) seem well separated<br />
from the cracked tiles (*). The objective is to find the two clusters.<br />
-8<br />
-8 -6 -4 -2 0 2<br />
log(intensity) 475 Hz<br />
1. Place two cluster centres (x) at random.<br />
2. Assign each data point (* and o) to the nearest cluster centre (x)<br />
2<br />
Tiles data: o = whole tiles, * = cracked tiles, x = centres<br />
2<br />
Tiles data: o = whole tiles, * = cracked tiles, x = centres<br />
1<br />
1<br />
0<br />
0<br />
log(intensity) 557 Hz<br />
-1<br />
-2<br />
-3<br />
-4<br />
-5<br />
log(intensity) 557 Hz<br />
-1<br />
-2<br />
-3<br />
-4<br />
-5<br />
-6<br />
-6<br />
-7<br />
-7<br />
-8<br />
-8 -6 -4 -2 0 2<br />
log(intensity) 475 Hz<br />
1. Compute the new centre of each class<br />
2. Move the crosses (x)<br />
-8<br />
-8 -6 -4 -2 0 2<br />
log(intensity) 475 Hz<br />
Iteration 2
log(intensity) 557 Hz<br />
2<br />
1<br />
0<br />
-1<br />
-2<br />
-3<br />
-4<br />
-5<br />
-6<br />
-7<br />
Tiles data: o = whole tiles, * = cracked tiles, x = centres<br />
-8<br />
-8 -6 -4 -2 0 2<br />
log(intensity) 475 Hz<br />
Iteration 3<br />
log(intensity) 557 Hz<br />
2<br />
1<br />
0<br />
-1<br />
-2<br />
-3<br />
-4<br />
-5<br />
-6<br />
-7<br />
Tiles data: o = whole tiles, * = cracked tiles, x = centres<br />
-8<br />
-8 -6 -4 -2 0 2<br />
log(intensity) 475 Hz<br />
Iteration 4 (then stop, because no visible change)<br />
Each data point belongs to the cluster defined by the nearest centre<br />
M =<br />
0.0000 1.0000<br />
0.0000 1.0000<br />
0.0000 1.0000<br />
0.0000 1.0000<br />
0.0000 1.0000<br />
1.0000 0.0000<br />
1.0000 0.0000<br />
1.0000 0.0000<br />
1.0000 0.0000<br />
1.0000 0.0000<br />
Membership matrix M<br />
m<br />
ik<br />
data point k cluster centre i<br />
⎧⎪<br />
1 if u −c ≤ u −c<br />
= ⎨<br />
⎪ ⎩0<br />
otherwise<br />
2<br />
k i k j<br />
distance<br />
2<br />
cluster centre j<br />
The membership matrix M:<br />
1. The last five data points (rows) belong to the first cluster (column)<br />
2. The first five data points (rows) belong to the second cluster (column)
c-partition<br />
Objective function<br />
All clusters C<br />
together fills the<br />
whole universe U<br />
Clusters do not<br />
overlap<br />
Minimise the total sum of<br />
all distances<br />
A cluster C is never<br />
empty and it is<br />
smaller than the<br />
whole universe U<br />
c<br />
∪<br />
i=<br />
1<br />
C = U<br />
C ∩ C = Ø<br />
i<br />
i<br />
Ø ⊂ C ⊂ U<br />
2 ≤ c ≤ K<br />
i<br />
j<br />
for all i ≠ j<br />
for all i<br />
There must be at least 2<br />
clusters in a c-partition and<br />
at most as many as the<br />
number of data points K<br />
J =<br />
c<br />
∑<br />
J<br />
=<br />
c<br />
⎛<br />
⎜<br />
⎝<br />
∑⎜<br />
∑<br />
i<br />
k<br />
i=<br />
1 i= 1 k , uk∈Ci<br />
u − c<br />
i<br />
2<br />
⎞<br />
⎟<br />
⎠<br />
<strong>Fuzzy</strong> c-<strong>means</strong><br />
One of the problems of the k-<strong>means</strong> algorithm is that it<br />
gives a hard partitioning of the data, that is to say that<br />
each point is attributed to one and only one cluster.<br />
But points on the edge of the cluster, or near another<br />
cluster, may not be as much in the cluster as points in<br />
the center of cluster.<br />
<strong>Fuzzy</strong> c-<strong>means</strong><br />
Therefore, in fuzzy clustering, each point does not pertain<br />
to a given cluster, but has a degree of belonging to a<br />
certain cluster, as in fuzzy logic. For each point x we have a<br />
coefficient giving the degree of being in the k-th cluster<br />
u k (x). Usually, the sum of those coefficients has to be one,<br />
so that u k (x) denotes a probability of belonging to a certain<br />
cluster:
<strong>Fuzzy</strong> c-<strong>means</strong><br />
<strong>Fuzzy</strong> c-<strong>means</strong><br />
The degree of being in a certain cluster is related to the<br />
inverse of the distance to the cluster<br />
With fuzzy c-<strong>means</strong>, the centroid of a cluster is computed as<br />
being the mean of all points, weighted by their degree of<br />
belonging to the cluster, that is:<br />
then the coefficients are normalized and fuzzyfied with a<br />
real parameter m > 1 so that their sum is 1. So :<br />
<strong>Fuzzy</strong> c-<strong>means</strong><br />
For m equal to 2, this is equivalent to normalising the<br />
coefficient linearly to make their sum 1. When m is close to<br />
1, then cluster center closest to the point is given much<br />
more weight than the others, and the algorithm is similar to<br />
k-<strong>means</strong>.<br />
<strong>Fuzzy</strong> c-<strong>means</strong><br />
The fuzzy c-<strong>means</strong> algorithm is greatly similar to the k-<br />
<strong>means</strong> algorithm :
<strong>Fuzzy</strong> c-<strong>means</strong><br />
•Choose a number of clusters<br />
•Assign randomly to each point coefficients for being in the<br />
clusters<br />
•Repeat until the algorithm has converged (that is, the<br />
coefficients' change between two iterations is no more than ε,<br />
the given sensitivity threshold) :<br />
•Compute the centroid for each cluster, using the formula<br />
above<br />
•For each point, compute its coefficients of being in the<br />
clusters, using the formula above<br />
<strong>Fuzzy</strong> C-<strong>means</strong><br />
uij is membership of sample i to custer j<br />
ck is centroid of custer i<br />
while changes in cluster Ck<br />
% compute new memberships<br />
for k=1,…,K do<br />
for i=1,…,N do<br />
ujk = f(xj – ck)<br />
end<br />
end<br />
% compute new cluster centroids<br />
for k=1,…,K do<br />
% weighted mean<br />
ck = SUMj jkxk xj /SUMj ujk<br />
end<br />
end<br />
<strong>Fuzzy</strong> c-<strong>means</strong><br />
<strong>Fuzzy</strong> c-<strong>means</strong> (FCM)<br />
The fuzzy c-<strong>means</strong> algorithm minimizes intra-cluster<br />
variance as well, but has the same problems as k-<strong>means</strong>,<br />
the minimum is local minimum, and the results depend on<br />
the initial choice of weights.<br />
log(intensity) 557 Hz<br />
2<br />
1<br />
0<br />
-1<br />
-2<br />
-3<br />
-4<br />
-5<br />
-6<br />
-7<br />
Tiles data: o = whole tiles, * = cracked tiles, x = centres<br />
-8<br />
-8 -6 -4 -2 0 2<br />
log(intensity) 475 Hz<br />
Each data point belongs to two clusters to different degrees
2<br />
Tiles data: o = whole tiles, * = cracked tiles, x = centres<br />
2<br />
Tiles data: o = whole tiles, * = cracked tiles, x = centres<br />
1<br />
1<br />
0<br />
0<br />
log(intensity) 557 Hz<br />
-1<br />
-2<br />
-3<br />
-4<br />
-5<br />
log(intensity) 557 Hz<br />
-1<br />
-2<br />
-3<br />
-4<br />
-5<br />
-6<br />
-6<br />
-7<br />
-7<br />
-8<br />
-8 -6 -4 -2 0 2<br />
log(intensity) 475 Hz<br />
-8<br />
-8 -6 -4 -2 0 2<br />
log(intensity) 475 Hz<br />
1. Place two cluster centres<br />
2. Assign a fuzzy membership to each data point depending on<br />
distance<br />
1. Compute the new centre of each class<br />
2. Move the crosses (x)<br />
2<br />
Tiles data: o = whole tiles, * = cracked tiles, x = centres<br />
2<br />
Tiles data: o = whole tiles, * = cracked tiles, x = centres<br />
1<br />
1<br />
0<br />
0<br />
log(intensity) 557 Hz<br />
-1<br />
-2<br />
-3<br />
-4<br />
-5<br />
log(intensity) 557 Hz<br />
-1<br />
-2<br />
-3<br />
-4<br />
-5<br />
-6<br />
-6<br />
-7<br />
-7<br />
-8<br />
-8 -6 -4 -2 0 2<br />
log(intensity) 475 Hz<br />
-8<br />
-8 -6 -4 -2 0 2<br />
log(intensity) 475 Hz<br />
Iteration 2<br />
Iteration 5
2<br />
Tiles data: o = whole tiles, * = cracked tiles, x = centres<br />
2<br />
Tiles data: o = whole tiles, * = cracked tiles, x = centres<br />
1<br />
1<br />
0<br />
0<br />
log(intensity) 557 Hz<br />
-1<br />
-2<br />
-3<br />
-4<br />
-5<br />
log(intensity) 557 Hz<br />
-1<br />
-2<br />
-3<br />
-4<br />
-5<br />
-6<br />
-6<br />
-7<br />
-7<br />
-8<br />
-8 -6 -4 -2 0 2<br />
log(intensity) 475 Hz<br />
-8<br />
-8 -6 -4 -2 0 2<br />
log(intensity) 475 Hz<br />
Iteration 10<br />
Iteration 13 (then stop, because no visible change)<br />
Each data point belongs to the two clusters to a degree<br />
M =<br />
0.0025 0.9975<br />
0.0091 0.9909<br />
0.0129 0.9871<br />
0.0001 0.9999<br />
0.0107 0.9893<br />
0.9393 0.0607<br />
0.9638 0.0362<br />
0.9574 0.0426<br />
0.9906 0.0094<br />
0.9807 0.0193<br />
<strong>Fuzzy</strong> membership matrix M<br />
Point k’s membership<br />
of cluster i<br />
m<br />
ik<br />
dik<br />
=<br />
c<br />
∑<br />
j=<br />
1<br />
k<br />
1<br />
⎛ ⎞<br />
⎜<br />
dik<br />
⎟<br />
⎝ d<br />
jk ⎠<br />
= u − c<br />
i<br />
2 /<br />
( q−1)<br />
Fuzziness<br />
exponent<br />
Distance from point k to<br />
current cluster centre i<br />
Distance from point k to<br />
other cluster centres j<br />
The membership matrix M:<br />
1. The last five data points (rows) belong mostly to the first cluster (column)<br />
2. The first five data points (rows) belong mostly to the second cluster (column)
<strong>Fuzzy</strong> membership matrix M<br />
m ik =<br />
2 / ( q−1<br />
⎛ ⎞<br />
)<br />
c<br />
dik<br />
=<br />
∑<br />
j=<br />
1<br />
=<br />
⎛ d<br />
⎜<br />
⎝ d<br />
d<br />
⎜<br />
⎝ d<br />
ik<br />
1k<br />
1<br />
1<br />
jk<br />
2 /( q−1) 2 /( q−1) 2 /( q−1)<br />
⎞<br />
⎟<br />
⎠<br />
⎟<br />
⎠<br />
⎛ d ⎞ ⎛<br />
ik<br />
d<br />
+ ⎜ ⎟ + +<br />
⎜<br />
⎝ d2k<br />
⎠ ⎝ d<br />
1<br />
2 /( q−1)<br />
dik<br />
1<br />
1<br />
+ + +<br />
2 /( q−1) 2 /( q−1) 2 /( q−1)<br />
1k<br />
d2k<br />
dck<br />
1<br />
ik<br />
ck<br />
⎞<br />
⎟<br />
⎠<br />
Gravitation to<br />
cluster i relative<br />
to total gravitation<br />
<strong>Fuzzy</strong> Membership<br />
Membership of test point<br />
1<br />
0.5<br />
o is with q = 1.1, * is with q = 2<br />
0<br />
1 2 3 4 5<br />
Data point<br />
Cluster centres<br />
<strong>Fuzzy</strong> c-partition<br />
Example: Classify cancer cells<br />
All clusters C together fill the<br />
whole universe U.<br />
Remark: The sum of<br />
memberships for a data point<br />
is 1, and the total for all<br />
points is K<br />
A cluster C is never<br />
empty and it is<br />
smaller than the<br />
whole universe U<br />
c<br />
∪<br />
i=<br />
1<br />
C = U<br />
C ∩ C = Ø<br />
i<br />
i<br />
Ø ⊂ C ⊂ U<br />
2 ≤ c ≤ K<br />
i<br />
j<br />
for all i ≠ j<br />
for all i<br />
Not valid: Clusters<br />
do overlap<br />
There must be at least 2<br />
clusters in a c-partition and<br />
at most as many as the<br />
number of data points K<br />
Normal smear<br />
Using a small brush, cotton stick, or wooden<br />
stick, a specimen is taken from the uterin cervix<br />
and smeared onto a thin, rectangular glass plate,<br />
a slide. The purpose of the smear screening is to<br />
diagnose pre-malignant cell changes before they<br />
progress to cancer. The smear is stained using<br />
the Papanicolau method, hence the name Pap<br />
smear. Different characteristics have different<br />
colours, easy to distinguish in a microscope. A<br />
cyto-technician performs the screening in a<br />
microscope. It is time consuming and prone to<br />
error, as each slide may contain up to 300.000<br />
cells.<br />
Severely dysplastic smear<br />
Dysplastic cells have undergone precancerous changes.<br />
They generally have longer and darker nuclei, and they<br />
have a tendency to cling together in large clusters. Mildly<br />
dysplastic cels have enlarged and bright nuclei.<br />
Moderately dysplastic cells have larger and darker<br />
nuclei. Severely dysplastic cells have large, dark, and<br />
often oddly shaped nuclei. The cytoplasm is dark, and it<br />
is relatively small.
Possible Features<br />
Classes are nonseparable<br />
• Nucleus and cytoplasm area<br />
• Nucleus and cyto brightness<br />
• Nucleus shortest and longest diameter<br />
• Cyto shortest and longest diameter<br />
• Nucleus and cyto perimeter<br />
• Nucleus and cyto no of maxima<br />
• (...)<br />
Hard Classifier (HCM)<br />
<strong>Fuzzy</strong> Classifier (FCM)<br />
moderate<br />
moderate<br />
Ok<br />
light<br />
Ok<br />
severe<br />
A cell is either one<br />
or the other class<br />
defined by a colour.<br />
Ok<br />
light<br />
Ok<br />
severe<br />
A cell can belong to<br />
several classes to a<br />
Degree, i.e., one column<br />
may have several colours.
Function approximation<br />
Approximation by fuzzy sets<br />
Output1<br />
1.5<br />
1<br />
0.5<br />
0<br />
-0.5<br />
2<br />
1<br />
0<br />
-1<br />
-2<br />
0 0.1 0.2 0.3 0.4 0.5 0.6 0.7 0.8 0.9 1<br />
-1<br />
-1.5<br />
0 0.1 0.2 0.3 0.4 0.5 0.6 0.7 0.8 0.9 1<br />
Input<br />
Curve fitting in a multi-dimensional space is also called function<br />
approximation. Learning is equivalent to finding a function that best<br />
fits the training data.<br />
1<br />
0.8<br />
0.6<br />
0.4<br />
0.2<br />
0<br />
0 0.1 0.2 0.3 0.4 0.5 0.6 0.7 0.8 0.9 1<br />
Procedure to find a model<br />
1. Acquire data<br />
2. Select structure<br />
3. Find clusters, generate model<br />
4. Validate model<br />
Conclusions<br />
• Compared to neural networks, fuzzy models can<br />
be interpreted by human beings<br />
• Applications: system identification, adaptive<br />
systems