Descriptive Stats and MATLAB support
Descriptive Stats and MATLAB support
Descriptive Stats and MATLAB support
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
BMED2803, Brani Vidakovic<br />
Sample <strong>and</strong> Its Properties II<br />
Multidimensional Samples<br />
1 Percentage of Body Fat from Simple Body Measurements<br />
We start wit a description of interesting data set from<br />
http://www.amstat.org/publications/jse/datasets/fat.txt<br />
http://www.amstat.org/publications/jse/datasets/fat.dat<br />
featured in Penrose, Nelson, <strong>and</strong> Fisher (1985). The data were generously supplied by Dr.<br />
A. Garth Fisher, Human Performance Research Center, Brigham Young University, Provo,<br />
UT, who gave permission to freely distribute the data <strong>and</strong> use them for non-commercial<br />
purposes.<br />
Percentage of body fat, age, weight, height, <strong>and</strong> ten body circumference measurements<br />
(e.g., abdomen) are recorded for 252 men. Body fat, a measure of health, is estimated<br />
through an underwater weighing technique. Fitting body fat to the other measurements<br />
using multiple regression provides a convenient way of estimating body fat for men using<br />
only a scale <strong>and</strong> a measuring tape.<br />
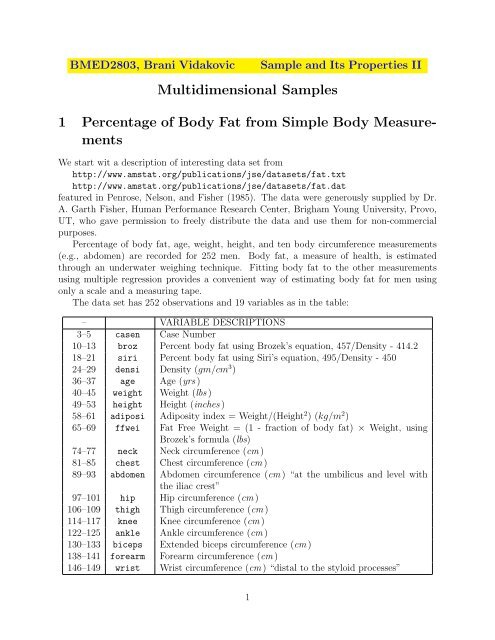
The data set has 252 observations <strong>and</strong> 19 variables as in the table:<br />
– VARIABLE DESCRIPTIONS<br />
3–5 casen Case Number<br />
10–13 broz Percent body fat using Brozek’s equation, 457/Density - 414.2<br />
18–21 siri Percent body fat using Siri’s equation, 495/Density - 450<br />
24–29 densi Density (gm/cm 3 )<br />
36–37 age Age (yrs )<br />
40–45 weight Weight (lbs )<br />
49–53 height Height (inches )<br />
58–61 adiposi Adiposity index = Weight/(Height 2 ) (kg/m 2 )<br />
65–69 ffwei Fat Free Weight = (1 - fraction of body fat) × Weight, using<br />
Brozek’s formula (lbs)<br />
74–77 neck Neck circumference (cm )<br />
81–85 chest Chest circumference (cm )<br />
89–93 abdomen Abdomen circumference (cm ) “at the umbilicus <strong>and</strong> level with<br />
the iliac crest”<br />
97–101 hip Hip circumference (cm )<br />
106–109 thigh Thigh circumference (cm )<br />
114–117 knee Knee circumference (cm )<br />
122–125 ankle Ankle circumference (cm )<br />
130–133 biceps Extended biceps circumference (cm )<br />
138–141 forearm Forearm circumference (cm )<br />
146–149 wrist Wrist circumference (cm ) “distal to the styloid processes”<br />
1
Remark: There are a few misrecordings. The body densities for cases 48, 76, <strong>and</strong> 96,<br />
for instance, each seem to have one digit in error as can be seen from the two body fat<br />
percentage values. Also note the presence of a man (case 42) over 200 pounds in weight who<br />
is less than 3 feet tall (the height should presumably be 69.5 inches, not 29.5 inches)! The<br />
percent body fat estimates are truncated to zero when negative (case 182).<br />
>> load(’C:\Courses\bmestatu\fat.dat’)<br />
>> casen = fat(:,1);<br />
>> broz = fat(:,2);<br />
>> siri = fat(:,3);<br />
>> densi = fat(:,4);<br />
>> age = fat(:,5);<br />
>> weight = fat(:,6);<br />
>> height = fat(:,7);<br />
>> adiposi = fat(:,8);<br />
>> ffwei = fat(:,9);<br />
>> neck = fat(:,10);<br />
>> chest = fat(:,11);<br />
>> abdomen = fat(:,12);<br />
>> hip = fat(:,13);<br />
>> thigh = fat(:,14);<br />
>> knee = fat(:,15);<br />
>> ankle = fat(:,16);<br />
>> biceps = fat(:,17);<br />
>> forearm = fat(:,18);<br />
>> wrist = fat(:,19);<br />
2 Covariances <strong>and</strong> Correlations<br />
Covariances <strong>and</strong> correlations are measuring relationship between paired data sets. Let X =<br />
(X 1 , X 2 , . . . , X n ) <strong>and</strong> Y = (Y 1 , Y 2 , . . . , Y n ) be two vectors so that components (X i , Y i ) are<br />
matched. We can think of observing pairs (X, Y) = ((X 1 , Y 1 ), . . . , (X n , Y n )). Covariance<br />
between paired samples X <strong>and</strong> Y is defined as<br />
Cov(X, Y) = 1<br />
n − 1<br />
n∑<br />
(X i − ¯X)(Y i − Ȳ ) = 1 [<br />
∑ n<br />
i=1<br />
n − 1<br />
X i Y i − n ¯XȲ<br />
i=1<br />
As in sample variances, the multiple 1/(n − 1) is sometimes replaced by 1/n.<br />
Pearson coefficient of correlation r between samples X <strong>and</strong> Y is their covariance normalized<br />
by their sample st<strong>and</strong>ard deviations,<br />
]<br />
.<br />
r = Corr(X, Y) =<br />
Cov(X, Y)<br />
s X s Y<br />
=<br />
∑ ni=1<br />
(X i − ¯X)(Y i − Ȳ )<br />
√ ∑ni=1<br />
(X i − ¯X) 2 × ∑ n<br />
i=1 (Y i − Ȳ )2 .<br />
2
where we canceled factor 1/(n − 1) in the numerator <strong>and</strong> denominator.<br />
This is Pearson’s coefficient of correlation <strong>and</strong> it measures the strength of linear relationship<br />
between samples X <strong>and</strong> Y.<br />
r is dimensionless, scale invariant, <strong>and</strong> bounded, −1 ≤ r ≤ 1. If r = 1 the relationship<br />
between X <strong>and</strong> Y is perfect linear, i.e., Y i = b 0 + b 1 X i , i = 1, . . . , n, for some constants b 0<br />
<strong>and</strong> b 1 > 0 If the linear relationship is valid but b 1 < 0, then r = −1. If r = 0, X <strong>and</strong> Y are<br />
uncorrelated, but not necessarily independent.<br />
Spearman coefficient of correlation operates on the ranks of the data. Ranks are ordinal<br />
numbers for the sorted data, the smallest observation has rank 1, next smallest 2, <strong>and</strong> so on.<br />
In case of ties, tied observations share an averaged rank. <strong>MATLAB</strong> program (home made,<br />
not part of <strong>MATLAB</strong>,<br />
ranks.m find the ranks for sequences <strong>and</strong> matrices of data. For example,<br />
>> x=r<strong>and</strong>(1,5); y=[x x(1)]<br />
y = 0.0153 0.7468 0.4451 0.9318 0.4660 0.0153<br />
>> ranks(y)<br />
ans = 1.5000 5.0000 3.0000 6.0000 4.0000 1.5000<br />
Notice that 1st <strong>and</strong> 6th observation match (by construction) <strong>and</strong> (that they are smallest.<br />
They both have ranks 1.5 as 1+2 , <strong>and</strong> the next larger observation has rank 3.<br />
2<br />
Now, Spearman’s coefficient of correlation r s is defined as Pearson coefficient of correlation<br />
among the ranks of X <strong>and</strong> Y. After some algebra,<br />
r s = 1 −<br />
6 ∑ d 2 i<br />
n(n 2 − 1) ,<br />
where d i = rank(X i ) − rank(Y i ).<br />
Kendall’s τ is measuring association of X <strong>and</strong> Y by selecting two out of n pairs <strong>and</strong><br />
selecting if the pairs are concordant or discordant. The numbers of concordant N c <strong>and</strong><br />
discordant N d pairs among ( )<br />
n<br />
2 = n(n − 1)/2 possible pairs are counted. Two pairs (Xi , Y i )<br />
<strong>and</strong> (X j , Y j ) are called concordant if (Y i −Y j )/(X i −X j ) > 0 <strong>and</strong> 1 is added to N c , discordant<br />
if (Y i − Y j )/(X i − X j ) < 0 <strong>and</strong> 1 is added to N d .<br />
If (Y i − Y j )/(X i − X j ) = 0 then 1/2 is added to both N c <strong>and</strong> N d <strong>and</strong> if X i = X j the<br />
concordance is not assessed.<br />
Then, Kendall’s τ is<br />
τ =<br />
N c − N d<br />
n(n − 1)/2 .<br />
>> X = [broz densi weight adiposi biceps];<br />
>> varNames = {’broz’; ’densi’; ’weight’; ’adiposi’; ’biceps’};<br />
>> varNames<br />
>> cov(X)<br />
>> corr(X)<br />
3
corr(X,’type’,’Spearman’)<br />
>> corr(X,’type’,’Kendall’)<br />
varNames =<br />
ans =<br />
’broz’<br />
’densi’<br />
’weight’<br />
’adiposi’<br />
’biceps’<br />
60.0758 -0.1458 139.6715 20.5847 11.5455<br />
-0.1458 0.0004 -0.3323 -0.0496 -0.0280<br />
139.6715 -0.3323 863.7227 95.1374 71.0711<br />
20.5847 -0.0496 95.1374 13.3087 8.2266<br />
11.5455 -0.0280 71.0711 8.2266 9.1281<br />
ans =<br />
ans =<br />
ans =<br />
1.0000 -0.9881 0.6132 0.7280 0.4930<br />
-0.9881 1.0000 -0.5941 -0.7147 -0.4871<br />
0.6132 -0.5941 1.0000 0.8874 0.8004<br />
0.7280 -0.7147 0.8874 1.0000 0.7464<br />
0.4930 -0.4871 0.8004 0.7464 1.0000<br />
1.0000 -0.9934 0.6126 0.7290 0.4926<br />
-0.9934 1.0000 -0.6014 -0.7224 -0.4892<br />
0.6126 -0.6014 1.0000 0.8696 0.7822<br />
0.7290 -0.7224 0.8696 1.0000 0.7663<br />
0.4926 -0.4892 0.7822 0.7663 1.0000<br />
1.0000 -0.9836 0.4356 0.5383 0.3434<br />
-0.9836 1.0000 -0.4297 -0.5309 -0.3389<br />
0.4356 -0.4297 1.0000 0.6880 0.5941<br />
0.5383 -0.5309 0.6880 1.0000 0.5805<br />
0.3434 -0.3389 0.5941 0.5805 1.0000<br />
4
Partial coefficient of correlation<br />
r X,Y ·Z =<br />
r XY − r XZ r Y Z<br />
√ √<br />
1 − rXZ<br />
2 1 − rY 2 Z<br />
>> figure(1)<br />
>> X = [broz densi weight adiposi biceps];<br />
>> varNames = {’broz’; ’densi’; ’weight’; ’adiposi’; ’biceps’};<br />
>> agegr = age > 55;<br />
>> gplotmatrix(X,[],agegr,[’b’,’r’],[’x’,’o’],[],’false’);<br />
>> text([.08 .24 .43 .66 .83], repmat(-.1,1,5), varNames, ’FontSize’,8);<br />
>> text(repmat(-.12,1,5), [.86 .62 .41 .25 .02], varNames, ’FontSize’,8,...<br />
’Rotation’,90);<br />
>> figure(2)<br />
>> %older = ismember(age,[55:max(age)])<br />
>> parallelcoords(X, ’group’, age>55, ...<br />
’st<strong>and</strong>ardize’,’on’, ’labels’,varNames)<br />
>> set(gcf,’color’,’white’);<br />
biceps<br />
adiposi weight<br />
densi<br />
broz<br />
40<br />
20<br />
1.1 0<br />
1.05<br />
350 1<br />
300<br />
250<br />
200<br />
150<br />
40<br />
20<br />
45<br />
40<br />
35<br />
30<br />
25<br />
0 20 40<br />
1 1.05 1.115020025030035020 40<br />
broz densi weight adiposi biceps<br />
(a)<br />
2530354045<br />
Coordinate Value<br />
8<br />
6<br />
4<br />
2<br />
0<br />
−2<br />
−4<br />
broz densi weight adiposi biceps<br />
(b)<br />
0<br />
1<br />
Figure 1: (a) gplotmatrix for broz, densi, weight, adiposi, <strong>and</strong> biceps ; (b)<br />
parallelcoords plot for X, by age > 55.<br />
5
figure(3)<br />
>> parallelcoords(X, ’group’, age>55, ...<br />
’st<strong>and</strong>ardize’,’on’, ’labels’,varNames,’quantile’,0.25)<br />
>> set(gcf,’color’,’white’);<br />
>> figure(4)<br />
>> <strong>and</strong>rewsplot(X, ’group’, age>55, ’st<strong>and</strong>ardize’,’on’)<br />
>> set(gcf,’color’,’white’);<br />
1.5<br />
1<br />
0<br />
1<br />
15<br />
10<br />
0<br />
1<br />
Coordinate Value<br />
0.5<br />
0<br />
−0.5<br />
f(t)<br />
5<br />
0<br />
−5<br />
−1<br />
−10<br />
−1.5<br />
broz densi weight adiposi biceps<br />
(a)<br />
−15<br />
0 0.2 0.4 0.6 0.8 1<br />
t<br />
(b)<br />
Figure 2: (a) X by age > 55 with quantiles; (b) <strong>and</strong>rewsplot for X by age > 55<br />
2.1 Visualizing Multivariate Data: Star Plots<br />
The data dimension d determines the number of rays exiting from the single point; the angle<br />
between two neighboring rays is 2π/d. The values of the data components are marked on the<br />
rays, ith component on the ray with angle (i − 1) ∗ 2π/d, i = 1, . . . , d. This defines spokes<br />
of the star. The star glyph connects the ends of the spokes. Thus, each spoke in a star<br />
represents one variable, <strong>and</strong> the spoke length is proportional to the value of that variable for<br />
that observation.<br />
>> figure(5)<br />
>> ind = find(age>67);<br />
>> strind = num2str(ind);<br />
>> h = glyphplot(X(ind,:), ’glyph’,’star’, ’varLabels’,varNames,...<br />
’obslabels’, strind);<br />
>> set(h(:,3),’FontSize’,8); set(gcf,’color’,’white’);<br />
6
2.2 Chernoff Faces<br />
The use of face representation is an interesting approach for a first look at multivariate<br />
data which is effective in revealing rather complex relations not always visible from simple<br />
correlations based on two-dimensional linear theories. It can be used to aid in cluster analysis,<br />
discrimination analysis <strong>and</strong> to detect substantial changes in time series. People grew up<br />
studying faces all the time. Small <strong>and</strong> barely measurable differences are easily detected<br />
<strong>and</strong> evoke emotional reactions from a long catalogue buried in memory. The human mind<br />
subconsciously operates as a high speed computer, filtering out insignificant phenomena <strong>and</strong><br />
focusing on the potentially important.<br />
The feature parameters are: Variable 1-Size of face; Variable 2-Forehead/jaw relative arc<br />
length; Variable 3-Shape of forehead; Variable 4-Shape of jaw; Variable 5-Width between<br />
eyes; Variable 6-Vertical position of eyes; Variables 7-13- features connected with location,<br />
separation, angle, shape <strong>and</strong> width of eyes <strong>and</strong> eyebrows; <strong>and</strong> so on.<br />
>> figure(6)<br />
>> ind = find(height > 74.5);<br />
>> strind = num2str(ind);<br />
>> h = glyphplot(X(ind,:), ’glyph’,’face’, ’varLabels’,varNames,...<br />
’obslabels’, strind);<br />
>> set(h(:,3),’FontSize’,10); set(gcf,’color’,’white’);<br />
78 79 84 85<br />
6 12 96<br />
87 246 247 248<br />
109 140 145<br />
249 250 251 252<br />
(a)<br />
156 192 194<br />
(b)<br />
Figure 3: (a) Star plots for X, (b) Chernoff faces plot for X.<br />
3 Arrythmia Example<br />
The data set arry.dat contains the following columns: 1. Arrhythmia presence/absence, 2.<br />
Age (years), 3. Aortic Cross Clamp Time, 4. Cardiopulminary Bypass Time, 5. ICU Time<br />
7
6. Avg Heart Rate, <strong>and</strong> 7. Left Ventricle Ejection Fraction.<br />
% ARRYTHMIA.M<br />
clear all<br />
close all<br />
disp(’Multivariate Data:Arrythmia Example’)<br />
%-----------------figure defaults<br />
lw = 3;<br />
set(0, ’DefaultAxesFontSize’, 16);<br />
fs = 14;<br />
msize = 5;<br />
r<strong>and</strong>n(’seed’,3)<br />
%-------------------------------------------------------------<br />
%1. Arrhythmia 2. Age<br />
%3. Aortic Cross Clamp Time 4. Cardiopulminary Bypass Time 5. ICU Time<br />
%6. Avg Heart Rate 7. Left Ventricle Ejection Fraction<br />
arry=[...<br />
1 68 64 126 20.25 85 81;...<br />
1 75 111 167 13.5 50 75;...<br />
1 69 63 94 12 74 62;...<br />
1 88 95 145 18.25 102.5 55;...<br />
1 75 82 132 13.25 90 78;...<br />
1 71 14 39 10.75 88.3 80;...<br />
1 72 80 124 12.5 88.5 56;...<br />
1 72 85 112 14 91 70;...<br />
1 72 97 128 14 89.5 40;...<br />
1 82 88 128 14 77.3 33;...<br />
1 83 49 105 14.5 79.7 65;...<br />
1 70 87 132 14.25 71.5 50;...<br />
1 65 98 137 15.33 89.4 38;...<br />
1 73 47 102 13.5 86.7 62;...<br />
1 79 108 150 16 86.7 55;...<br />
1 70 83 122 18 74 50;...<br />
1 73 81 120 14 68.8 55;...<br />
1 64 0 0 14 86.2 55;...<br />
1 81 79 157 19.5 98 80;...<br />
1 71 81 123 22 97 70;...<br />
1 77 107 149 17 75.8 46;...<br />
1 53 120 137 17.5 96.5 25;...<br />
1 66 86 109 20.75 72.8 42.5;...<br />
1 76 61 133 17 82.5 55;...<br />
1 75 97 149 12.67 93.8 80;...<br />
1 74 70 147 13.42 95 80;...<br />
1 75 72 156 8 82 70;...<br />
1 66 98 137 15 90 35;...<br />
0 48 102 169 18.5 76.2 59;...<br />
0 47 81 111 12.25 70 66;...<br />
0 62 76 126 18.5 101.4 56;...<br />
0 71 82 130 13.62 97.6 67;...<br />
0 48 132 224 20.5 103.2 44;...<br />
0 85 91 124 18.17 76.3 82;...<br />
0 54 44 89 12 87.4 65;...<br />
8
0 69 124 210 15 81.6 60;...<br />
0 45 105 157 14.75 96 43;...<br />
0 53 67 119 2 94.6 64;...<br />
0 51 23 42 17.25 74 73;...<br />
0 70 93 148 19.25 73.2 46;...<br />
0 69 55 109 18.5 109.3 60;...<br />
0 76 67 101 12.25 97.4 60;...<br />
0 61 105 185 22.5 98.4 35;...<br />
0 69 153 233 22.5 66.4 60;...<br />
0 50 84 113 19.5 97.3 40;...<br />
0 70 107 142 20 82.3 59;...<br />
0 55 116 140 13.5 70 55;...<br />
0 72 100 148 18 80.4 53;...<br />
0 76 77 180 23 93.2 50;...<br />
0 77 63 109 13 87.6 39;...<br />
0 65 78 117 16 59.2 68;...<br />
0 57 67 103 15 79.2 70;...<br />
0 70 107 137 19 66.8 60;...<br />
0 69 193 487 20.75 104 60;...<br />
0 74 90 169 14.5 94.2 50;...<br />
0 62 71 123 13.5 66.2 65;...<br />
0 67 72 105 19.25 76 60;...<br />
0 64 139 196 21 77.2 18;...<br />
0 59 55 78 12.25 88 55;...<br />
0 44 47 77 21 107 60;...<br />
0 51 77 138 19 99 40;...<br />
0 48 113 120 15 82.2 60;...<br />
0 74 0 0 12.5 79.3 63;...<br />
0 66 80 131 20.75 88.5 60;...<br />
0 68 94 158 18 88 30;...<br />
0 81 90 121 14 81.4 60;...<br />
0 70 87 120 21 81 51;...<br />
0 76 72 108 19.5 91.8 50;...<br />
0 66 112 163 17.75 89.8 50;...<br />
0 61 82 129 14 76.3 60;...<br />
0 73 46 80 20.75 80.6 50;...<br />
0 44 87 175 15.25 97.4 65;...<br />
0 68 102 137 13.25 71.8 71;...<br />
0 60 99 143 18.5 85 72;...<br />
0 73 61 91 17.5 85.6 37;...<br />
0 69 50 98 11.67 84 55;...<br />
0 64 63 118 13.25 97.8 40;...<br />
0 44 75 147 18.583 111.8 60;...<br />
0 71 51 83 16.75 102 59;...<br />
0 73 68 122 17.833 78.2 50;...<br />
0 55 44 78 11.5 94.8 40];<br />
%---------------------------------------------------------------------<br />
arryth= arry(:,1);<br />
age=arry(:,2);<br />
aoclati = arry(:,3);<br />
cbt=arry(:,4);<br />
icu = arry(:,5);<br />
9
avehrate=arry(:,6);<br />
lventr=arry(:,7);<br />
%------------------<br />
X = [age, aoclati, cbt, icu, avehrate, lventr];<br />
varNames = {’age’,’aoclati’,’cbt’,’icu’,’avehrate’,’lventr’};<br />
% covariances <strong>and</strong> correlations<br />
cov(X)<br />
corr(X)<br />
corr(X,’type’,’Spearman’)<br />
corr(X,’type’,’Kendall’)<br />
corr(arryth, icu)<br />
figure(1)<br />
agegr = age > 60;<br />
gplotmatrix(X,[],agegr,[’b’,’r’],[’x’,’o’],[],’false’);<br />
print -depsc ’C:\Brani\Courses\bmestatu\Fall2007\arry1.eps’<br />
figure(2)<br />
older = ismember(age,[70:max(age)]);<br />
parallelcoords(X, ’group’, older, ...<br />
’st<strong>and</strong>ardize’,’on’, ’labels’,varNames)<br />
set(gcf,’color’,’white’);<br />
print -depsc ’C:\Brani\Courses\bmestatu\Fall2007\arry2.eps’<br />
figure(3)<br />
parallelcoords(X, ’group’, icu>16, ...<br />
’st<strong>and</strong>ardize’,’on’, ’labels’,varNames,’quantile’,0.25)<br />
set(gcf,’color’,’white’);<br />
print -depsc ’C:\Brani\Courses\bmestatu\Fall2007\arry3.eps’<br />
figure(4)<br />
<strong>and</strong>rewsplot(X, ’group’, age>60, ’st<strong>and</strong>ardize’,’on’)<br />
set(gcf,’color’,’white’);<br />
print -depsc ’C:\Brani\Courses\bmestatu\Fall2007\arry4.eps’<br />
figure(5)<br />
ind = find(age>67);<br />
strind = num2str(ind);<br />
h = glyphplot(X(ind,:), ’glyph’,’star’, ’varLabels’,varNames,...<br />
’obslabels’, strind);<br />
set(h(:,3),’FontSize’,8); set(gcf,’color’,’white’);<br />
print -depsc ’C:\Brani\Courses\bmestatu\Fall2007\arry5.eps’<br />
figure(6)<br />
ind = find(avehrate > 95);<br />
strind = num2str(ind);<br />
h = glyphplot(X(ind,:), ’glyph’,’face’, ’varLabels’,varNames,...<br />
’obslabels’, strind);<br />
set(h(:,3),’FontSize’,10); set(gcf,’color’,’white’);<br />
print -depsc ’C:\Brani\Courses\bmestatu\Fall2007\arry6.eps’<br />
10
4 Exercises <strong>and</strong> Examples<br />
Mr Joseph Bentley, the owner of the pharmacy store wants to remove the Coke vending machine st<strong>and</strong>ing in<br />
front of his store because he believes the vending machine influences the number of errors the store employees<br />
make. More precisely, the more Coke is sold the more errors in his store is made. He provided the following<br />
data:<br />
errors 10 6 21 23 9 15 17 8<br />
coke 112 100 220 250 100 200 160 100<br />
<strong>and</strong> indeed, the coefficient of correlation is 0.9552. But we can explain this correlation because we have<br />
complete data including the lurking variable – the number of people that passes by his store on the street,<br />
errors 10 6 21 23 9 15 17 8<br />
coke 112 100 220 250 100 200 160 100<br />
people 10000 6000 17000 20000 9000 15000 14000 8000<br />
<strong>and</strong> all the sudden the correlation disappears.<br />
>> mederrors = [10, 6, 21, 23, 9, 15, 17, 8];<br />
>> coke = [112, 100, 220, 250, 100, 200, 160, 100];<br />
>> people = [10000, 6000, 17000, 20000, 9000, 15000, 14000, 8000];<br />
>> corr(mederrors’, coke’)<br />
ans = 0.9552<br />
>> corr(mederrors’, people’)<br />
ans = 0.9845<br />
>> corr(coke’, people’)<br />
ans = 0.9735<br />
>> (0.9552 - 0.9845 * 0.9735)/(sqrt(1-0.9845^2)*sqrt(1-0.9735^2))<br />
ans = -0.0801<br />
5 Refrences<br />
Penrose, K., Nelson, A., <strong>and</strong> Fisher, A. (1985), ”Generalized Body Composition Prediction Equation for Men<br />
Using Simple Measurement Techniques” (abstract), Medicine <strong>and</strong> Science in Sports <strong>and</strong> Exercise, 17(2), 189.<br />
11