A combined discreteâcontinuous model describing the lag phase of ...
A combined discreteâcontinuous model describing the lag phase of ...
A combined discreteâcontinuous model describing the lag phase of ...
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
174 R.C. McKellar, K. Knight / International Journal <strong>of</strong> Food Microbiology 54 (2000) 171 –180<br />
Table 1<br />
Kinetic parameters for Listeria monocytogenes determined using<br />
a<br />
<strong>the</strong> Bioscreen<br />
Trial t m t S.D. % Growth<br />
d L L<br />
(n520)<br />
A 20.35 1.04 5.86 0.783 60<br />
B 21.32 0.876 4.12 0.814 65<br />
a 21<br />
t<br />
d, Time to detection (h); m, specific growth rate (h ); t<br />
L,<br />
mean individual cell <strong>lag</strong> <strong>phase</strong> duration (h); S.D.<br />
L, standard<br />
deviation <strong>of</strong> <strong>the</strong> mean individual <strong>lag</strong> <strong>phase</strong> duration; % growth,<br />
percent <strong>of</strong> wells showing growth.<br />
single cell per well can be calculated from a Poisson<br />
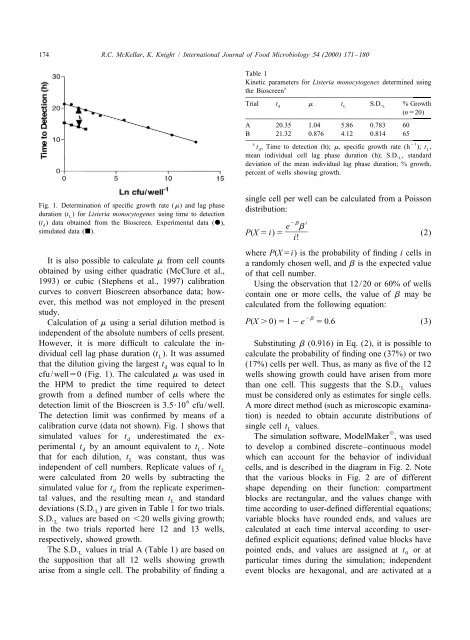
Fig. 1. Determination <strong>of</strong> specific growth rate (m) and <strong>lag</strong> <strong>phase</strong><br />
duration (t<br />
L) for Listeria monocytogenes using time to detection<br />
distribution:<br />
(t ) data obtained from <strong>the</strong> Bioscreen. Experimental data (d),<br />
2b i<br />
d<br />
e b<br />
simulated data (j). P(X 5 i) 5 ]]<br />
i!<br />
(2)<br />
where P(X5i) is <strong>the</strong> probability <strong>of</strong> finding i cells in<br />
It is also possible to calculate m from cell counts a randomly chosen well, and b is <strong>the</strong> expected value<br />
obtained by using ei<strong>the</strong>r quadratic (McClure et al., <strong>of</strong> that cell number.<br />
1993) or cubic (Stephens et al., 1997) calibration Using <strong>the</strong> observation that 12/20 or 60% <strong>of</strong> wells<br />
curves to convert Bioscreen absorbance data; how- contain one or more cells, <strong>the</strong> value <strong>of</strong> b may be<br />
ever, this method was not employed in <strong>the</strong> present calculated from <strong>the</strong> following equation:<br />
study.<br />
Calculation <strong>of</strong> m using a serial dilution method is<br />
independent <strong>of</strong> <strong>the</strong> absolute numbers <strong>of</strong> cells present.<br />
2b<br />
P(X . 0) 5 1 2 e 5 0.6 (3)<br />
However, it is more difficult to calculate <strong>the</strong> in- Substituting b (0.916) in Eq. (2), it is possible to<br />
dividual cell <strong>lag</strong> <strong>phase</strong> duration (t<br />
L). It was assumed calculate <strong>the</strong> probability <strong>of</strong> finding one (37%) or two<br />
that <strong>the</strong> dilution giving <strong>the</strong> largest td<br />
was equal to ln (17%) cells per well. Thus, as many as five <strong>of</strong> <strong>the</strong> 12<br />
cfu/well50 (Fig. 1). The calculated m was used in wells showing growth could have arisen from more<br />
<strong>the</strong> HPM to predict <strong>the</strong> time required to detect than one cell. This suggests that <strong>the</strong> S.D.<br />
L<br />
values<br />
growth from a defined number <strong>of</strong> cells where <strong>the</strong> must be considered only as estimates for single cells.<br />
6<br />
detection limit <strong>of</strong> <strong>the</strong> Bioscreen is 3.510 cfu/well. A more direct method (such as microscopic examina-<br />
The detection limit was confirmed by means <strong>of</strong> a tion) is needed to obtain accurate distributions <strong>of</strong><br />
calibration curve (data not shown). Fig. 1 shows that single cell tL<br />
values.<br />
©<br />
simulated values for t underestimated <strong>the</strong> ex- The simulation s<strong>of</strong>tware, ModelMaker , was used<br />
d<br />
perimental td<br />
by an amount equivalent to t<br />
L. Note to develop a <strong>combined</strong> discrete–continuous <strong>model</strong><br />
that for each dilution, tL<br />
was constant, thus was which can account for <strong>the</strong> behavior <strong>of</strong> individual<br />
independent <strong>of</strong> cell numbers. Replicate values <strong>of</strong> t cells, and is described in <strong>the</strong> diagram in Fig. 2. Note<br />
L<br />
were calculated from 20 wells by subtracting <strong>the</strong> that <strong>the</strong> various blocks in Fig. 2 are <strong>of</strong> different<br />
simulated value for td<br />
from <strong>the</strong> replicate experimen- shape depending on <strong>the</strong>ir function: compartment<br />
tal values, and <strong>the</strong> resulting mean tL<br />
and standard blocks are rectangular, and <strong>the</strong> values change with<br />
deviations (S.D. ) are given in Table 1 for two trials. time according to user-defined differential equations;<br />
L<br />
S.D.<br />
L<br />
values are based on ,20 wells giving growth; variable blocks have rounded ends, and values are<br />
in <strong>the</strong> two trials reported here 12 and 13 wells, calculated at each time interval according to userrespectively,<br />
showed growth.<br />
defined explicit equations; defined value blocks have<br />
The S.D.<br />
L<br />
values in trial A (Table 1) are based on pointed ends, and values are assigned at t0<br />
or at<br />
<strong>the</strong> supposition that all 12 wells showing growth particular times during <strong>the</strong> simulation; independent<br />
arise from a single cell. The probability <strong>of</strong> finding a<br />
event blocks are hexagonal, and are activated at a