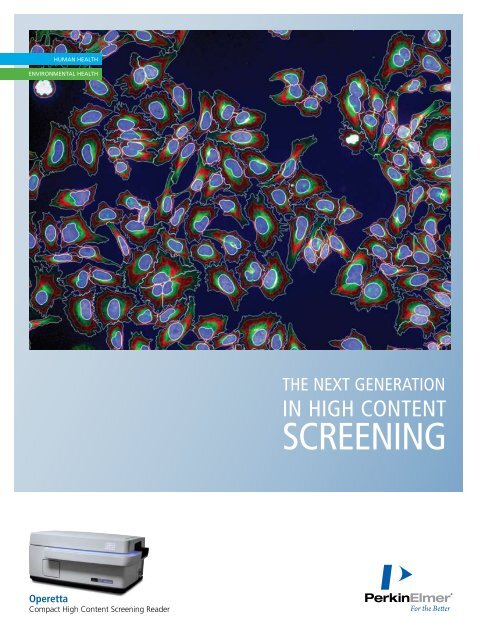
Operetta The Next Generation in High Content Screening
Operetta The Next Generation in High Content Screening
Operetta The Next Generation in High Content Screening
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
<strong>Operetta</strong><br />
Compact <strong>High</strong> <strong>Content</strong> Screen<strong>in</strong>g Reader<br />
the next generation<br />
<strong>in</strong> high content<br />
screen<strong>in</strong>g
<strong>Operetta</strong><br />
<strong>Operetta</strong> –<br />
the high content<br />
screen<strong>in</strong>g solution<br />
for every lab<br />
Offer<strong>in</strong>g a new bench-top<br />
approach to <strong>High</strong> <strong>Content</strong><br />
Screen<strong>in</strong>g (HCS), the<br />
<strong>Operetta</strong> is easy to use,<br />
allow<strong>in</strong>g researchers who are new to HCS to use preconfigured<br />
applications and quickly become productive. User-configurable<br />
build<strong>in</strong>g blocks enable more experienced users to develop their<br />
own assays, or modify exist<strong>in</strong>g applications. <strong>The</strong> compact design<br />
fits any size laboratory to provide a versatile solution that suits<br />
the needs of every HCS facility.<br />
Benefit<strong>in</strong>g from many years of experience creat<strong>in</strong>g outstand<strong>in</strong>g<br />
solutions for HCS, Perk<strong>in</strong>Elmer br<strong>in</strong>gs new, state-of-the-art<br />
technology to HCS research. Our <strong>in</strong>-depth knowledge of the<br />
technical requirements for successful high content imag<strong>in</strong>g is<br />
complemented by our understand<strong>in</strong>g of researchers' assays and<br />
applications. Provid<strong>in</strong>g a total solution for beg<strong>in</strong>ners and experts<br />
alike, the <strong>Operetta</strong> <strong>in</strong>corporates both eng<strong>in</strong>eer<strong>in</strong>g excellence and<br />
a complete, easy-to-use software <strong>in</strong>terface.<br />
Key Features<br />
• Easy-to-use solution for <strong>High</strong> <strong>Content</strong> Screen<strong>in</strong>g –<br />
<strong>in</strong>tuitive user <strong>in</strong>terface, short learn<strong>in</strong>g curve and<br />
onl<strong>in</strong>e user guides enable fast results<br />
• Exceptional image quality<br />
• Powerful image analysis <strong>in</strong>cludes automatic parameter<br />
optimization for ease-of-use<br />
• Database with keyword browser enables search<strong>in</strong>g<br />
by user, application, date, screen and compound<br />
<strong>Operetta</strong> Application Guide<br />
To help new HCS users, we provide an easy to follow<br />
guide to analyz<strong>in</strong>g your results. This step-by-step<br />
handbook will help you through the process, whether<br />
you are us<strong>in</strong>g a ready-made solution or design<strong>in</strong>g a<br />
new image analysis sequence from the build<strong>in</strong>g blocks<br />
provided. <strong>The</strong> guide <strong>in</strong>cludes general considerations,<br />
recommendations and common pitfalls, so that<br />
you can proceed with confidence and produce<br />
results quickly.<br />
<strong>The</strong> <strong>Operetta</strong> has a compact design that will fit <strong>in</strong>to any laboratory.<br />
<strong>The</strong> large work<strong>in</strong>g area beneath the lid allows easy access to samples.<br />
2
<strong>Operetta</strong> Hardware<br />
Designed to provide the best functionality <strong>in</strong> the smallest<br />
possible footpr<strong>in</strong>t, the <strong>Operetta</strong> has a sleek profile and status<br />
<strong>in</strong>dicator illum<strong>in</strong>ation that allows you to track the progress<br />
of your experiment with ease. Lift<strong>in</strong>g the lid on the <strong>Operetta</strong><br />
reveals a large work<strong>in</strong>g area allow<strong>in</strong>g easy access to the sample<br />
area, while the eight position emission filter wheel is conveniently<br />
housed so that filters can be changed quickly and easily.<br />
Barcod<strong>in</strong>g prevents filter set-up errors, and maximum flexibility<br />
is achieved with up to 15 emission filter comb<strong>in</strong>ations.<br />
All standard objectives are barcoded for convenience, provid<strong>in</strong>g<br />
a wide range of choices for any plate type. <strong>High</strong> numerical<br />
apertures enable optimal resolution and sensitivity. Long work<strong>in</strong>g<br />
distance objectives allow the use of any plate type. <strong>The</strong><br />
custom-designed light path permits a large field of view, greatly<br />
<strong>in</strong>creas<strong>in</strong>g the number of cells visualized without compromis<strong>in</strong>g<br />
resolution and throughput.<br />
Available with brightfield, widefield, and confocal options, the<br />
system can cater for your application needs both now and <strong>in</strong><br />
the future. <strong>High</strong> resolution is provided with a Nipkow disk to<br />
enhance axial resolution and to suppress out of focus light.<br />
Xenon<br />
lamp<br />
Transmission light<br />
Attenuator<br />
Sample<br />
Excitation filter<br />
wheel<br />
Objective turret<br />
Shutter<br />
Shutter<br />
Emission filter<br />
wheel<br />
Camera<br />
Dichroic mirror<br />
Optional confocal light<br />
path, p<strong>in</strong>hole disk<br />
Autofocus<br />
<strong>The</strong> optical path of the <strong>Operetta</strong><br />
www.perk<strong>in</strong>elmer.com/operetta<br />
3
<strong>Operetta</strong> – hardware and software <strong>in</strong> complete harmony<br />
Provid<strong>in</strong>g shared, remote and dedicated access, the Harmony software is designed to streaml<strong>in</strong>e<br />
and simplify the HCS process. With its <strong>in</strong>tuitive user <strong>in</strong>terface that leads you through your<br />
HCS experiment, Harmony suggests next steps where appropriate, simplify<strong>in</strong>g the process<br />
for both new and experienced HCS users.<br />
Harmony <strong>in</strong>cludes all the tools you need to develop an assay, automate the hardware, analyze your data, and store and retrieve<br />
the results. Remote access to the <strong>Operetta</strong> is available via your office computer and local network for analyz<strong>in</strong>g and view<strong>in</strong>g<br />
your results.<br />
Harmony for Flexible Acquisition<br />
Acquisition controls are displayed with<strong>in</strong> a unified w<strong>in</strong>dow with predef<strong>in</strong>ed solutions or options to create protocols to your own<br />
requirements. Multiple acquisition options are available, <strong>in</strong>clud<strong>in</strong>g channels, fields per well and z-sections. All imag<strong>in</strong>g parameter<br />
metadata, such as dyes / filters, objectives, plate type, fields, z-stack or exposure, are embedded <strong>in</strong>to the experiment files and<br />
stored <strong>in</strong> the database for future reference.<br />
Harmony for Rapid, Accurate Analysis<br />
Ready-Made Solutions<br />
A full range of ready-made solutions for frequently used image analysis tasks and assays is pre-<strong>in</strong>stalled for your convenience,<br />
mak<strong>in</strong>g it ideal for new users. Harmony still provides the flexibility to change and modify your experiments us<strong>in</strong>g image<br />
analysis build<strong>in</strong>g blocks.<br />
Harmony Ready-Made Solution Applications (examples)<br />
Cell Count<strong>in</strong>g or Nuclei Count<strong>in</strong>g<br />
Live / Dead Cell Count<strong>in</strong>g<br />
Quantification of Marker <strong>in</strong> Nucleus<br />
Quantification of Marker <strong>in</strong> Cytoplasm<br />
Quantification of Marker <strong>in</strong> Membrane<br />
Cytosol to Nucleus Translocation<br />
Cytosol to Membrane Translocation<br />
Fluorescence Redistribution – Cytoskeleton<br />
proliferation, cytotoxicity<br />
cell viability: live / dead cell count<br />
Prote<strong>in</strong> expression <strong>in</strong> nucleus (e.g. caspase-3) • Cell cycle: S-Phase quantification (BrdU, EdU)<br />
Prote<strong>in</strong> expression <strong>in</strong> cytosol • Mitochondrial mass • Loss of mitochondrial membrane potential<br />
Phospholipidosis • Hepatosteatosis • Cell cycle: Transition of G2 to M-phase (cycl<strong>in</strong> B1)<br />
Prote<strong>in</strong> expression <strong>in</strong> plasma membrane • Signall<strong>in</strong>g pathways: Membrane ruffl<strong>in</strong>g<br />
Signall<strong>in</strong>g pathways: Transcription factor activation<br />
Signall<strong>in</strong>g pathways: Cytoplasm to membrane translocation<br />
Cytoskeletal rearrangement<br />
Spot Analysis Peroxisome quantification • Vesicle formation • GPCR activation: arrest<strong>in</strong> recruitment (e.g. Transfluor ® )<br />
Nuclear Spots<br />
Nuclear Analysis – Nuclear Shr<strong>in</strong>k<strong>in</strong>g<br />
Nuclear Fragmentation<br />
Nuclear Classification – DNA <strong>Content</strong><br />
Cell Shape – Cell Round<strong>in</strong>g<br />
fluorescence In Situ Hybridization (FISH)<br />
Nuclear swell<strong>in</strong>g and shr<strong>in</strong>k<strong>in</strong>g<br />
apoptosis / Nuclear fragmentation<br />
Cell cycle: DNA content<br />
Cell spread<strong>in</strong>g, cell detachment, cell round<strong>in</strong>g<br />
Mitotic Index Cell cycle: Mitotic <strong>in</strong>dex (phosphohistone H3)<br />
Cell Cycle Classification<br />
Simultaneous phosphohistone H3, EdU, DNA content and nuclear morphology<br />
Harmony <strong>in</strong>cludes preconfigured image analysis sequences for common applications. Known as ready-made solutions, they are ideal for new users of HCS.<br />
4
Custom-Designed Solutions<br />
Harmony provides a complete range of build<strong>in</strong>g blocks to enable simplified custom image analysis. Each clearly def<strong>in</strong>ed block has<br />
primary tun<strong>in</strong>g parameters with optimized values that can be automatically or manually adjusted with real-time visual feedback to<br />
facilitate the set-up process.<br />
Harmony Build<strong>in</strong>g Block Description<br />
F<strong>in</strong>d Nuclei<br />
F<strong>in</strong>d Cytoplasm<br />
F<strong>in</strong>d Cells<br />
F<strong>in</strong>d Spots<br />
F<strong>in</strong>d Image Region<br />
Select Cell Region<br />
Select Region<br />
Calculate Intensity Properties<br />
Calculate Morphology Properties<br />
Calculate Properties<br />
Select Population<br />
identifies and counts cellular nuclei based on a nuclear sta<strong>in</strong><br />
Identifies cell bodies / cytoplasmic area when nuclei have been identified before<br />
F<strong>in</strong>ds whole cells based on fluorescence <strong>in</strong>tensity e.g. for whole cell fluorescence assays<br />
Identifies fluorescent objects <strong>in</strong> cells of similar size, such as endosomes, vesicles, peroxisomes<br />
Segments the entire image based on fluorescence <strong>in</strong>tensity<br />
Def<strong>in</strong>es a region <strong>in</strong> the cytoplasm or nucleus, based on nucleus and cytoplasm identification<br />
Def<strong>in</strong>es a sub-region<br />
calculates fluorescence <strong>in</strong>tensity of regions / objects, provides mean,<br />
median, standard deviation, CV, max, m<strong>in</strong>, contrast values<br />
Calculates morphology properties of regions / objects, such as area, length, width,<br />
axis, ratio, roundness<br />
Allows any comb<strong>in</strong>ation of <strong>in</strong>tensity and morphology properties<br />
Def<strong>in</strong>es sub-populations of cells<br />
Harmony for Data Storage and Retrieval<br />
A scalable database is <strong>in</strong>cluded for management of all images, experiment <strong>in</strong>formation and data analysis. Metadata <strong>in</strong>formation,<br />
such as exposure parameters, analysis method and users, is embedded <strong>in</strong>to all data files, while a keyword browser facilitates<br />
search<strong>in</strong>g us<strong>in</strong>g those keywords for your projects. A sortable database <strong>in</strong>formation tree allows quick and easy access to any<br />
desired <strong>in</strong>formation.<br />
Automatic parameter adjustment helps you f<strong>in</strong>d the best sett<strong>in</strong>gs quickly.<br />
F<strong>in</strong>d images, metadata and results easily <strong>in</strong> the Harmony database.<br />
www.perk<strong>in</strong>elmer.com/operetta<br />
5
<strong>Operetta</strong> — compact <strong>in</strong>strument provid<strong>in</strong>g the complete solution<br />
<strong>Operetta</strong> Hardware<br />
Light sources<br />
• Xenon fibre optic light source, 300 W,<br />
360 – 640 nm cont<strong>in</strong>uous spectrum<br />
• LED light source for transmission mode<br />
Excitation and emission<br />
• Fully automated eight position excitation filter wheel<br />
• Fully automated eight position emission filter wheel<br />
• 15 emission filters available, barcoded and user exchangeable<br />
Camera<br />
• 14-bit Peltier-cooled camera<br />
• CCD chip size 1360 x 1024 pixel, 6.45 µm square<br />
Objectives<br />
• 4 position objective turret for automated change<br />
• 10x high NA and long WD<br />
• 20x high NA and long WD<br />
• 40x long WD<br />
• 60x long WD<br />
• 100x long WD<br />
Scan table<br />
• <strong>High</strong> resolution scann<strong>in</strong>g stage<br />
(150 nm mechanical stage x-y accuracy)<br />
Harmony Software<br />
Image acquisition<br />
• Computer controlled excitation power<br />
• Laser-based fully automated and fast autofocus<strong>in</strong>g<br />
• Preconfigured plate formats (96, 384)<br />
• User def<strong>in</strong>ed plate formats; free def<strong>in</strong>ition also possible<br />
• Assay layout creation with arbitrary layers of <strong>in</strong>formation<br />
• Multiple image fields with<strong>in</strong> wells<br />
• Z-stack measurements<br />
• Fully network compatible<br />
Image analysis<br />
• Fully automated cell segmentation with automatic<br />
or manual parameter tun<strong>in</strong>g<br />
• Ready-made turnkey applications<br />
• Easy and <strong>in</strong>tuitive creation and modification of analysis<br />
sequences<br />
Data management<br />
• Scalable database<br />
• Search via sortable tree<br />
• Stores all metadata automatically<br />
(assay layout, <strong>in</strong>strument sett<strong>in</strong>g, analysis results)<br />
• Stores additional experiment <strong>in</strong>formation<br />
(user def<strong>in</strong>ed keywords, annotations)<br />
• Allows for image storage on a remote server<br />
Computer<br />
Processor<br />
Memory<br />
Hard Drive<br />
Network <strong>in</strong>terface<br />
Intel ® Core 2 Quad 2,83 GHz<br />
4 GB<br />
500 GB (RAID 1, 2 x 500 GB)<br />
Operat<strong>in</strong>g system W<strong>in</strong>dows Vista ®<br />
Gigabit Ethernet<br />
24" flat screen<br />
Physical Dimensions<br />
Width: 33" (83 cm)<br />
Depth: 21" (53 cm)<br />
Height: 16" (40 cm)<br />
<strong>Operetta</strong> status <strong>in</strong>dicator illum<strong>in</strong>ation helps monitor<br />
the progress of experiments.<br />
6
<strong>Operetta</strong> – use the network solution to maximize results<br />
By us<strong>in</strong>g the local network, you can perform tasks such as experiment design and view<strong>in</strong>g<br />
results, so that the <strong>Operetta</strong> is free for other researchers to use, maximiz<strong>in</strong>g the productivity<br />
of your system.<br />
Def<strong>in</strong>e assay layout<br />
Set up on the <strong>Operetta</strong> system<br />
or transfer to the <strong>Operetta</strong><br />
from your office computer via<br />
the network when required.<br />
Optional steps<br />
Develop/modify image analysis,<br />
re-analyze images<br />
Select measurement<br />
protocol<br />
Perform on the <strong>Operetta</strong> system, or<br />
transfer to your office computer via<br />
the network.<br />
Select image analysis<br />
solution<br />
Automated measurement<br />
Onl<strong>in</strong>e image analysis<br />
View results<br />
View on the <strong>Operetta</strong> system,<br />
or transfer to your office<br />
computer via the network.<br />
www.perk<strong>in</strong>elmer.com/operetta<br />
7
Perk<strong>in</strong>Elmer support to suit your needs<br />
Perk<strong>in</strong>Elmer is committed to provid<strong>in</strong>g outstand<strong>in</strong>g after-sales support us<strong>in</strong>g our network of local, highly tra<strong>in</strong>ed service eng<strong>in</strong>eers. As<br />
an <strong>Operetta</strong> user you can choose the level of support that you require, from ma<strong>in</strong>tenance agreements for hardware or software, right<br />
up to full 24 hour response.<br />
Perk<strong>in</strong>Elmer’s <strong>High</strong> <strong>Content</strong> Screen<strong>in</strong>g product solutions<br />
Opera FLIM <strong>High</strong> <strong>Content</strong><br />
Screen<strong>in</strong>g System<br />
<strong>The</strong> Opera FLIM is a time-doma<strong>in</strong><br />
fluorescence lifetime imag<strong>in</strong>g reader<br />
with ultra-high speed data acquisition.<br />
Opera QEHS <strong>High</strong> <strong>Content</strong><br />
Screen<strong>in</strong>g System<br />
<strong>The</strong> Opera is the premier confocal<br />
microplate imag<strong>in</strong>g reader provid<strong>in</strong>g<br />
solutions for fully automated simultaneous<br />
multi-color high speed and high resolution<br />
screen<strong>in</strong>g.<br />
Acapella Image Analysis Software<br />
Acapella is a flexible and powerful<br />
image data process<strong>in</strong>g system for a<br />
broad range of applications.<br />
Opera LX <strong>High</strong> <strong>Content</strong><br />
Screen<strong>in</strong>g System<br />
<strong>The</strong> Opera LX confocal microplate<br />
imag<strong>in</strong>g reader is an extendable starter<br />
configuration for three color applications.<br />
<strong>Operetta</strong> Compact <strong>High</strong><br />
<strong>Content</strong> Screen<strong>in</strong>g System<br />
<strong>The</strong> <strong>Operetta</strong> is an easy-to-use<br />
bench-top high content screen<strong>in</strong>g<br />
<strong>in</strong>strument. Includes an <strong>in</strong>tuitive<br />
user <strong>in</strong>terface, multiple wavelength<br />
fluorescence and brightfield imag<strong>in</strong>g.<br />
Columbus Data Management<br />
and Analysis Software<br />
Columbus sets the <strong>in</strong>dustry standard for<br />
high volume image data management<br />
provid<strong>in</strong>g image storage, retrieval,<br />
visualization and analysis.<br />
Disclaimer: A user license to the <strong>The</strong>rmoFisher Scientific (Cellomics) HCS patent portfolio is <strong>in</strong>cluded with each <strong>Operetta</strong> and Harmony product.<br />
For the use of the assays described additional licenses may be required.<br />
Perk<strong>in</strong>Elmer, Inc.<br />
940 W<strong>in</strong>ter Street<br />
Waltham, MA 02451 USA<br />
P: (800) 762-4000 or<br />
(+1) 203-925-4602<br />
www.perk<strong>in</strong>elmer.com<br />
For a complete list<strong>in</strong>g of our global offices, visit www.perk<strong>in</strong>elmer.com/ContactUs<br />
Copyright ©2009, Perk<strong>in</strong>Elmer, Inc. All rights reserved. Perk<strong>in</strong>Elmer ® is a registered trademark of Perk<strong>in</strong>Elmer, Inc. All other trademarks are the property of their respective owners.<br />
008451B_01<br />
Pr<strong>in</strong>ted <strong>in</strong> USA