DICTYOTALES, PHAEOPHYCEAE - Phycology Research Group ...
DICTYOTALES, PHAEOPHYCEAE - Phycology Research Group ...
DICTYOTALES, PHAEOPHYCEAE - Phycology Research Group ...
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
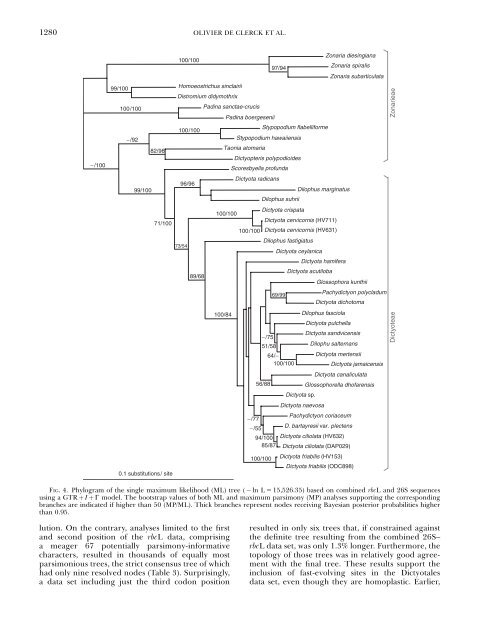
1280OLIVIER DE CLERCK ET AL.−/10099/100100/100−/9282/9899/10071/1000.1 substitutions/ site100/100Homoeostrichus sinclairii97/94Zonaria diesingianaZonaria spiralisZonaria subarticulataDistromium didymothrixPadina sanctae-crucisPadina boergeseniiStypopodium flabelliforme100/100Stypopodium hawaiiensisTaonia atomariaDictyopteris polypodioidesScoresbyella profundaDictyota radicans96/96Dilophus marginatusDilophus suhriiDictyota crispata100/100Dictyota cervicornis (HV711)100/100 Dictyota cervicornis (HV631)Dilophus fastigiatusDictyota ceylanicaDictyota hamiferaDictyota acutiloba89/68Glossophora kunthii69/99Pachydictyon polycladumDictyota dichotoma100/84Dilophus fasciolaDictyota pulchellaDictyota sandvicensis−/7551/58Dilophu salternans64/−Dictyota mertensii100/100Dictyota jamaicensisDictyota canaliculata56/88Glossophorella dhofarensisDictyota sp.Dictyota naevosaPachydictyon coriaceum−/77−/55D. bartayresii var. plectens94/100 Dictyota ciliolata (HV632)85/87 Dictyota ciliolata (DAP029)100/100 Dictyota friabilis (HV153)Dictyota friabilis (ODC898)ZonarieaeDictyoteaeFIG. 4. Phylogram of the single maximum likelihood (ML) tree ( ln L 5 15,526.35) based on combined rbcL and 26S sequencesusing a GTR þ I þ G model. The bootstrap values of both ML and maximum parsimony (MP) analyses supporting the correspondingbranches are indicated if higher than 50 (MP/ML). Thick branches represent nodes receiving Bayesian posterior probabilities higherthan 0.95.lution. On the contrary, analyses limited to the firstand second position of the rbcL data, comprisinga meager 67 potentially parsimony-informativecharacters, resulted in thousands of equally mostparsimonious trees, the strict consensus tree of whichhad only nine resolved nodes (Table 3). Surprisingly,a data set including just the third codon positionresulted in only six trees that, if constrained againstthe definite tree resulting from the combined 26S–rbcL dataset,wasonly1.3%longer.Furthermore,thetopology of those trees was in relatively good agreementwith the final tree. These results support theinclusion of fast-evolving sites in the Dictyotalesdata set, even though they are homoplastic. Earlier,