T cell Receptor
T cell Receptor
T cell Receptor
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
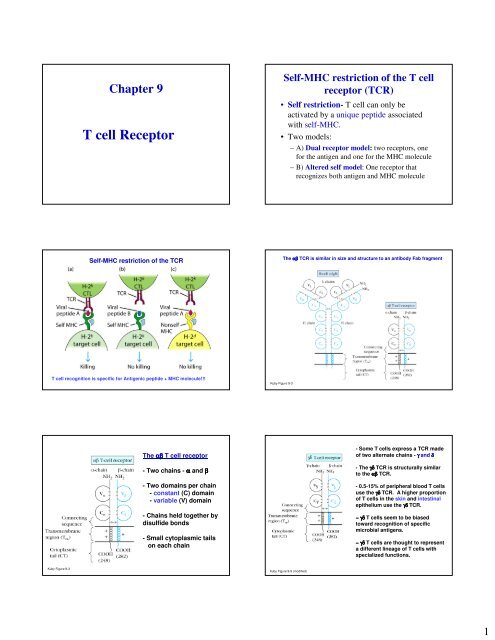
Chapter 9T <strong>cell</strong> <strong>Receptor</strong>Self-MHC restriction of the T <strong>cell</strong>receptor (TCR)• Self restriction- T <strong>cell</strong> can only beactivated by a unique peptide associatedwith self-MHC.• Two models:– A) Dual receptor model: two receptors, onefor the antigen and one for the MHC molecule– B) Altered self model: One receptor thatrecognizes both antigen and MHC moleculeSelf-MHC restriction of the TCRThe αβ TCR is similar in size and structure to an antibody Fab fragmentT <strong>cell</strong> recognition is specific for Antigenic peptide + MHC molecule!!!Kuby Figure 9-3The αβ T <strong>cell</strong> receptor- Two chains - α and β- Two domains per chain- constant (C) domain- variable (V) domain- Chains held together bydisulfide bonds- Small cytoplasmic tailson each chain- Some T <strong>cell</strong>s express a TCR madeof two alternate chains - γ and δ- The γδ TCR is structurally similarto the αβ TCR.- 0.5-15% of peripheral blood T <strong>cell</strong>suse the γδ TCR. A higher proportionof T <strong>cell</strong>s in the skin and intestinalepithelium use the γδ TCR.− γδ T <strong>cell</strong>s seem to be biasedtoward recognition of specificmicrobial antigens.− γδ T <strong>cell</strong>s are thought to representa different lineage of T <strong>cell</strong>s withspecialized functions.Kuby Figure 9-3Kuby Figure 9-9 (modified)1
Table 9.1 Comparison of TCRαβ T <strong>cell</strong>sγδ T <strong>cell</strong>s• % CD3 + 90-99% 1-10%• TCR V gene Large Smallin germline• CD4/CD8CD4 60%
1, rearrangement1, rearrangement2, RNA transcript3, Post-transcriptional modifications4, mature mRNA5, polypeptide6, Post-translational modificationsRearrangement of TCR genes• TCR Genes also composed of V, D, J and C genesegments• Genes are located in different chromosomes• The β and δ chains contain D segments (like Ig Heavychains!) while the α and γ chains do not.• α and γ chains - VJ rearrangement only• β and δ chains - DJ and then V-DJ rearrangement• Segments of the δ chain are embedded within thesegments encoding the α chain• When the α chain rearranges, δ segments are deleted• T <strong>cell</strong>s express only αβ or γδ TCR• Rearrangement involves RAG-1 and RAG-2 and TdT• Rearrangement is governed by the one turn-two turnruleGeneration of antibody diversity1. Multiple germline V, D and J gene segments2. Combinatorial V-J and V-D-J joining3. Somatic hypermutation4. Junctional flexibility5. P-nucleotide addition6. N-nucleotide additionGeneration of TCR diversity-Varying number of D segments in the delta (and beta) chain, why?(arrangement of RSS sequences differs from that in Ig loci to allowthis)Generation of TCR diversity2) N-region nucleotide addition, occurs in all chainsAntibody12 bp23 bp1.6 x 10 11 VS 3 x 10 7Note: Increased diversity in TCR!3
MAJOR DIFFERENCESBETWEEN TCR AND Ig GENES• Somatic hyper-mutation (affinity maturation)- During an antibody response, mutations accumulate at arapid rate in the VDJ gene segments encoding the BCR.- Thus, as an immune response proceeds, the affinity of theantibody produced (i.e. its ability to bind to the antigen)increases.• Alternative joining of D segments (β, δ)• N-nucleotide addition to both chains***1.6 x 10 11 VS 3 x 10 7Properties of Ig and TCR GenesIg TCRMany VDJs, few Cs yes yesVDJ rearrangement yes yesV-regions form antigen yes yesrecognition siteSomatic hypermutation yes noProperties of Ig and TCRProteinsIg TCRTransmembrane forms yes yesSecreted forms yes noIsotypes with different yes no*functionsValency 2 1The TCR complex includes CD3 - 3 heterodimers: γε, εδ and ζζ- 1) TCR is not expressed without CD3. It is required to bringTCR to surface- 2) All chains of CD3 possess ITAM motifs. (Immunoreceptortyrosine-based activation motif) Signal TransductionTCR <strong>Receptor</strong> Complex- CD3RECOGNITIONSIGNAL TRANSDUCTION4
WHY ACCESSORY MOLECULES?1) Due to low affinity of TCR with peptide MHCcomplex2) Provide:- Adhesion, Activation and Co-stimulation- Some show increased expression in response tocytokinesRECAP:-The BCR consists of IgM or IgD plus Ig-α/Ig-βheterodimers. The Ig binds the antigen while theIg-α/Ig-β heterodimers are involved in activation ofthe B <strong>cell</strong>.- The TCR consists of either the α/β chains or the γ/δchains plus CD3. The αβ or γδ chains bind theantigen while CD3 is involved in activation of the T <strong>cell</strong>.*Accessory Molecules Involvedin Cell-Cell InteractionsInteractions of Th Cell and APCCD4+ T <strong>cell</strong> CD-2 LFA-1 TCR CD28 CD45RIL-1IL-6TNF-alphaIL-12IL-15peptideCD4TNF-betaIFN-gammaGM-CSFIL-4APCLFA-3ICAM-1Class IIMHCB7-1/B7-2(CD80/CD86CD22Interactions of Tc and Target CellCD8+ T <strong>cell</strong> LFA-1 TCRCD2 CD45RCD8peptideT-<strong>cell</strong> Accessory molecules• CD4 and CD8 are co-receptors because theyrecognize the peptide-MHC complex• CD8 recognizes the α3 MHC-I domain; whileCD4 interacts with α2 MHC-II domain• Both CD4 and CD8 act in signal transduction• OTHERTarget<strong>cell</strong>ICAM-1Class IMHCLFA-3CD225
Accessory Molecules Involvedin Cell-Cell InteractionsCell Adhesion:T CellCD2- (LFA-2)LFA-1Ligand on APCLFA-3ICAM-1, ICAM-2LFA = Leukocyte Function-associatedAntigenICAM = InterCellular Adhesion MoleculeCostimulatory Molecules• Molecules on T <strong>cell</strong> and 2 nd <strong>cell</strong> that engageto deliver 2 nd signal required for activation ofT <strong>cell</strong>• Most important co-stimulatory molecules:T <strong>cell</strong> Ligand on 2 nd <strong>cell</strong>CD28 B7-1 (CD80), B7-2 (CD86)CTLA-4 B7-1 (CD80), B7-2 (CD86)CD45RCD22CD4/CD8 MHC-I/IIT <strong>cell</strong>APCAlloreactive T <strong>cell</strong>s• Allogeneic – genetically different individual of thesame species (humans!!)• MHC molecules are alloantigens: CD4 T <strong>cell</strong>s arealloreactive to MHC-II alloantigens and CD8 T<strong>cell</strong>s are alloreactive to MHC-I alloantigens• Direct and Indirect recognition of alloantigens: a)nonself MHC moleucle on foreign <strong>cell</strong> recognizedin its native form; b) peptides from allogeneicMHC after processing are prpresented by self-MHC molecules• The End6