The University of Tennessee Health Science Center
The University of Tennessee Health Science Center
The University of Tennessee Health Science Center
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
For more information about the <strong>University</strong> <strong>of</strong> <strong>Tennessee</strong> <strong>Health</strong> <strong>Science</strong> <strong>Center</strong>, visit www.utmem.edu.<br />
This brochure is a joint project <strong>of</strong> <strong>The</strong> <strong>University</strong> <strong>of</strong> <strong>Tennessee</strong> <strong>Health</strong> <strong>Science</strong>s <strong>Center</strong> and <strong>The</strong> <strong>University</strong> <strong>of</strong> Memphis.<br />
This brochure was organized by the UT <strong>Health</strong> <strong>Science</strong> <strong>Center</strong> Offi ce <strong>of</strong> the Vice Chancellor for Research and<br />
produced by the UTHSC Communications and Marketing Department.<br />
<strong>The</strong> <strong>University</strong> <strong>of</strong> <strong>Tennessee</strong> is an EEO/AA/ Title VI/Title IX/Section 504/ADA/ADEA institution in the provision <strong>of</strong> its education<br />
and employment programs and services.<br />
E070401-009-07
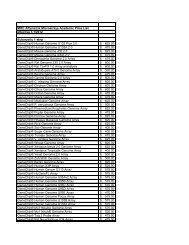
Table <strong>of</strong> Contents<br />
Biomedical Instrumentation . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 2<br />
Comparative Medicine and Laboratory Animal Care Unit . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 3<br />
Drosophila Core . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 3<br />
Flow Cytometry and Cell Sorting Facility . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 4<br />
Genomics and Bioinformatics Cores . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 6<br />
Imtek MicroCat/SPECT System . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 7<br />
<strong>University</strong> <strong>of</strong> Memphis<br />
Integrated Microscopy <strong>Center</strong>, Departments <strong>of</strong> Physics, Biomedical Engineering and<br />
W. Harry Feinstone <strong>Center</strong> for Genomic Research and Bioinformatics . . . . . . . . . . . . . . . . . . . . . 8<br />
Laser Capture Microdissection . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 11<br />
Mass Spectrometry Core Laboratory . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 12<br />
Molecular Resource <strong>Center</strong> . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 13<br />
Neuroscience Imaging . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 15<br />
Public Parking at UTHSC . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 16<br />
1
2<br />
Biomedical Instrumentation<br />
S<br />
ince 1948, the Biomedical Instrumentation (BMI)<br />
team at the <strong>University</strong> <strong>of</strong> <strong>Tennessee</strong> campus in<br />
Memphis has provided a diverse array <strong>of</strong> services<br />
to customers both inside and outside <strong>of</strong> the<br />
<strong>University</strong>. Over the years, the BMI team has served thousands<br />
<strong>of</strong> customers in the public and private<br />
sector throughout the Mid-South with<br />
services that include: Computer Repair,<br />
Electronic Fabrication, Mechanical<br />
Design and Fabrication, Equipment<br />
Design and Repair, Rehabilitation<br />
Engineering Services, and Microscope<br />
Repair.<br />
BMI has built a solid reputation for<br />
timeliness, expertise, value and<br />
reasonable pricing. <strong>The</strong> team produces<br />
high quality work at costs that are<br />
consistently below most market rates.<br />
In addition, their customer service<br />
orientation and in-depth knowledge allow<br />
them to generate innovative solutions that<br />
serve the clients’ best interests.<br />
Computer Repair<br />
� Certifi ed as authorized service<br />
center for Dell, Gateway and Apple<br />
computers<br />
� Services other brands <strong>of</strong> computers,<br />
printers and peripherals<br />
� Performs system, disk drive and<br />
memory upgrades<br />
� Executes new equipment setup and<br />
checkout and can relocate existing<br />
equipment<br />
Mechanical Design and Fabrication<br />
� CAD/CAM<br />
� Can custom design work for<br />
researchers<br />
� Experienced as a small-scale producer<br />
<strong>of</strong> parts<br />
� Has 5th Axis milling capabilities<br />
� Offers complete machine shop<br />
services<br />
� Three CNC milling machines<br />
� Performs welding, brazing and<br />
soldering – aluminum, stainless steel<br />
� Utilizes plastic welding equipment for PVC and<br />
polyethylene<br />
� Provides relocation <strong>of</strong> delicate and expensive equipment<br />
Laboratory Equipment Repair<br />
� Repairs most manufacturers’<br />
equipment<br />
� Provides consultation, calibration<br />
and verifi cation <strong>of</strong> system<br />
parameters<br />
� Offers preventive maintenance<br />
� Performs new equipment setup and<br />
checkout<br />
Rehabilitation Engineering<br />
Services<br />
� Installs computer hardware<br />
adaptations<br />
� Performs s<strong>of</strong>tware adaptations<br />
� Executes computer repair<br />
� Provides custom fabrication services<br />
� Offers experienced counselors to<br />
assist clients with technology<br />
consultations<br />
Microscope Repair<br />
� Provides cleaning service and repair<br />
� Maintains an available stock <strong>of</strong><br />
replacement lamps<br />
<strong>The</strong> BMI team’s expertise, gleaned from<br />
years <strong>of</strong> practical experience, result in<br />
real savings for customers. <strong>The</strong> team can<br />
extend the life <strong>of</strong> existing equipment<br />
through repair rather than replacement.<br />
For additional information contact:<br />
Biomedical Instrumentation<br />
822 Beale Street<br />
Beale Building, Room 141<br />
Memphis, TN 38163<br />
(901) 448-5652<br />
(901) 448-5337 (fax)<br />
instrument@utmem.edu<br />
www.utmem.edu/BMI
<strong>The</strong> Department <strong>of</strong> Comparative Medicine and Laboratory<br />
Animal Care Unit provides suitable housing facilities, animal<br />
husbandry services, and research technical services for<br />
animals used in biomedical research. <strong>The</strong> program is overseen<br />
by veterinarians who are board certifi ed in laboratory animal<br />
medicine who provide veterinary and diagnostic services,<br />
personnel training and expertise in laboratory animal research,<br />
surgery, and model development.<br />
<strong>The</strong> Molecular Resource <strong>Center</strong> is proud to announce the<br />
Drosophila melanogaster transgenic core facility (DTC) in the<br />
department <strong>of</strong> Neurology at the <strong>University</strong> <strong>of</strong> <strong>Tennessee</strong> <strong>Health</strong><br />
<strong>Science</strong> <strong>Center</strong>. This facility will be the fi rst <strong>of</strong> its kind in the<br />
Memphis area and will further enhance genetic approaches<br />
to translational research at UTHSC. <strong>The</strong> purpose <strong>of</strong> the<br />
Drosophila Transgenic Core (DTC) is to encourage<br />
investigator’s to utilize the genetic screening tools in Drosophila<br />
to quickly identify genetic pathways or individual genes related<br />
to their disease gene <strong>of</strong> interest. <strong>The</strong> primary tools <strong>of</strong> the<br />
DTC will be the construction <strong>of</strong> transgenic Drosophila lines<br />
containing the investigator’s gene <strong>of</strong> interest under the<br />
control <strong>of</strong> the yeast upstream activator sequence (UAS). We<br />
can assist these investigators in generating visible phenotypes in<br />
the eye, wing, larvae or other tissues using various GAL4<br />
drivers maintained in the lab. Using the Drosophila deletion kits<br />
Comparative Medicine and Laboratory<br />
Animal Care Unit<br />
For additional information contact:<br />
Timothy Mandrell, DVM, DACLAM<br />
Pr<strong>of</strong>essor and Chair<br />
Comparative Medicine<br />
(901) 448-5656<br />
tmandrell@utmem.edu<br />
www.utmem.edu/research/LACU<br />
Drosophila Core<br />
(both Bloomington and DrosDel collections) we will help<br />
train postdocs or graduate students from the investigator’s<br />
laboratory to screen for potential suppressors or enhancers <strong>of</strong><br />
the phenotypes generated. Additional projects can also be<br />
performed on a case-by-case basis, such as the creation <strong>of</strong> lines<br />
for proteomic analysis, single neuron mutation analysis or<br />
analysis <strong>of</strong> mutants affecting the nervous system.<br />
For additional information contact:<br />
Larry Reiter, Ph.D.<br />
Neurology<br />
Room 431 Johnson<br />
(901) 448-2635 � (901) 448-6199 (alternate)<br />
lreiter@utmem.edu<br />
http://curlyo.utmem.edu<br />
3
4<br />
Flow Cytometry and Cell Sorting Facility<br />
T<br />
he Laboratory provides access to state-<strong>of</strong>-the-art<br />
analytical fl ow cytometry and cell sorting for<br />
UTHSC and Memphis area researchers. <strong>The</strong><br />
instruments available are a BD Biosciences<br />
FACSAria Cell Sorter, a BD Biosciences LSR II Flow Cytometer,<br />
and a BD Biosciences FACSCalibur Flow Cytometer.<br />
Who<br />
<strong>The</strong> Administrative/Faculty Director <strong>of</strong> the Laboratory is<br />
Dr. Tony Marion, Department <strong>of</strong> Molecular <strong>Science</strong>s. <strong>The</strong><br />
Director <strong>of</strong> Operations is Dan Rosson, Ph.D. Dr. Rosson is the<br />
contact person for access and scheduling <strong>of</strong> services provided<br />
by the laboratory.<br />
Where<br />
<strong>The</strong> facilities are located in Rm. 214,<br />
Molecular <strong>Science</strong>s Building, 858<br />
Madison and Rm. 146, <strong>The</strong> Cancer<br />
Research Building, 19 South Manassas,<br />
both within the UTHSC campus. <strong>The</strong><br />
LSR II fl ow cytometer and FACSAria cell<br />
sorter are in MSB214. <strong>The</strong> FACSCalibur<br />
fl ow cytometer is in Rm. 146, Cancer<br />
Research Building.<br />
How<br />
Individuals wishing to schedule<br />
training, use <strong>of</strong> either <strong>of</strong> the fl ow<br />
cytometers, operator-assisted fl ow<br />
cytometry, or cell sorting should contact<br />
Dr. Rosson, MSB214, 448-4279,<br />
drosson@utmem.edu. Researchers may<br />
choose to collect fl ow cytometry data<br />
themselves on either the FACSCalibur<br />
or LSR II. Only operator-assisted fl ow<br />
cytometry and cell sorting is accessible with the FACSAria.<br />
Users <strong>of</strong> the FACSCalibur and the LSR II must fi rst complete<br />
a two-hour training course with Dr. Rosson or provide<br />
documented evidence <strong>of</strong> training and complete familiarity with<br />
the relevant instrument’s operation. Training, access to the fl ow<br />
cytometers, and scheduling for cell sorting or operator-assisted<br />
analysis requires an account number to which all charges may<br />
be submitted for payment.<br />
When<br />
Cell sorting and fl ow cytometry services and facilities are<br />
available Monday through Friday, 9:00 a.m. – 5:00 p.m. and at<br />
other times by special request.<br />
Flow cytometric analyses available:<br />
� Differentiation <strong>of</strong> relative cell size and complexity for<br />
eukaryotic and prokaryotic cells<br />
� Simultaneous detection and quantifi cation <strong>of</strong> expression<br />
<strong>of</strong> up to 11 cell surface and/or intracellular molecules with<br />
fl urophore-labeled antibodies<br />
� Ca2+ fl ux measurement among cell subpopulations<br />
defi ned by multiple cell-surface markers<br />
� Detection and quantifi cation <strong>of</strong> GFP-labeled protein<br />
expression<br />
� Ca2+ fl ux during cell signaling among cell subpopulations<br />
defi ned by cell surface markers<br />
� DNA quantifi cation and cell cycle analysis<br />
� Detection and quantifi cation <strong>of</strong><br />
apoptosis<br />
� High Throughput Screening (HTS)<br />
<strong>of</strong> cells or fl urophore-labeled beads<br />
for the detection <strong>of</strong> cytokines or<br />
cell signaling phosphoproteins, i.e.<br />
BD Cytometric Bead Array® (CBA)<br />
Cell sorting services available:<br />
� High-speed sorting <strong>of</strong> cells<br />
identifi ed with up to 12 cell surface<br />
and/or intracellular fl uorescent<br />
markers into four or fewer defi ned<br />
subpopulations<br />
� Detection and high-speed sorting<br />
<strong>of</strong> bacteria or eukaryotic subcellular<br />
particles<br />
� Sorting into microwell plates<br />
� Single-cell deposition into microwell<br />
plates or onto microscope slides<br />
Flow Cytometry Facility<br />
<strong>The</strong> fl ow cytometry facility is contained and maintained within<br />
the Department <strong>of</strong> Molecular <strong>Science</strong>s, MSB214, and the<br />
Cancer Research Building, Rm. 146. <strong>The</strong> facility includes two<br />
analytical fl ow cytometers and a cell sorter. <strong>The</strong> facility<br />
functions as a core laboratory accessible to all PIs and research<br />
laboratories at UTHSC as well as other institutions and<br />
companies in the Memphis area and is operated on a fee-forservice<br />
basis. <strong>The</strong> facility is directed by Dr. Tony Marion,<br />
Pr<strong>of</strong>essor, Department <strong>of</strong> Molecular <strong>Science</strong>s and has a fulltime,<br />
highly trained operator, Dr. Dan Rosson. Dr. Rosson<br />
has many years <strong>of</strong> training and expertise in cell biology,<br />
particularly cancer cell biology, and fl ow cytometry. Both Drs.
Rosson and Marion are available to all users for help in<br />
designing and implementing protocols to implement fl ow<br />
cytometry and cell sorting within their research projects.<br />
Cell Sorter:<br />
FACSAria Special Order System<br />
(BD Biosciences)<br />
<strong>The</strong> cell sorter is equipped with four lasers with 12 fl uorescence<br />
detectors, in addition to FSC and SSC PMTs. <strong>The</strong> 0-100 mW<br />
488 nm blue diode laser is equipped with fi ve fl uorescence<br />
detectors and a SSC detector in an octagon confi guration.<br />
This laser is fi ltered for detection <strong>of</strong> FITC, PE, PerCP-Cy5.5,<br />
PE-TxRd, and PE-Cy7. <strong>The</strong> 30 mW 638 nm red diode laser<br />
is equipped with three fl uorescence detectors in a trigon<br />
confi guration, and is fi ltered for detection <strong>of</strong> APC, APC-Cy7,<br />
and Alexa700. <strong>The</strong> 50 mW<br />
405 nm violet diode laser<br />
has two fl uorescence<br />
detectors in a trigon<br />
confi guration, and is fi ltered<br />
for detection <strong>of</strong> Pacifi c Blue/<br />
Cascade Blue/Alexa405 and<br />
AmCyan/Pacifi c Orange/<br />
Alexa430. <strong>The</strong> 20 mW<br />
355 nm solid-state UV<br />
laser has two fl uorescence<br />
detectors in an octagon<br />
confi guration, and has<br />
upgrade capacity for Qdots.<br />
Detection <strong>of</strong> Hoechst,<br />
Indo-1 (Ca-bound and<br />
unbound), and DAPI can<br />
be accomplished using<br />
this laser. FSC detection<br />
is selectable between PMT<br />
(for increased sensitivity<br />
and detection <strong>of</strong> subcellular<br />
particles and bacteria < 0.2 micrometer) and diode (for<br />
eukaryotic cells). <strong>The</strong> instrument has two-and four-way sort<br />
capacity (into tubes or microtubes) with sort rates <strong>of</strong> 22,000<br />
events/sec in purity sort mode with predicted recovery <strong>of</strong> 80%<br />
and 98% purity. This sort rate allows for sorting the entire<br />
mononuclear cell population from a single mouse spleen in<br />
approximately 1 hour.<br />
<strong>The</strong> unit is also specially equipped with:<br />
� temperature controlled sample injection and cell<br />
collection chambers<br />
� biosafety level-2 aerosol containment (for sorting<br />
infectious samples)<br />
� automated cell deposition (including single cell) into<br />
microwell plates or onto microscope slides.<br />
Flow Cytometer:<br />
LSR II Special Option Flow Cytometer<br />
(BD Biosciences)<br />
<strong>The</strong> LSR II fl ow cytometer is equipped with four lasers with<br />
eleven fl uorescence detectors, in addition to FSC and SSC<br />
PMTs. <strong>The</strong> 22 mW 488 nm blue laser is equipped with four<br />
fl uorescence detectors and a SSC PMT, in an octagon<br />
confi guration, and is fi ltered for detection <strong>of</strong> FITC, PE,<br />
PerCP-Cy5.5, PE-TxRd, and PE-Cy7. <strong>The</strong> 20 mW 638 nm red<br />
laser has three fl uorescence detectors (in a trigon confi guration)<br />
which are capable <strong>of</strong> detecting APC, APC-Cy7, and Alexa700.<br />
<strong>The</strong> 22 mW 405 nm violet laser has two fl uorescence detectors<br />
(in a trigon confi guration), that have capacity to detect Pacifi c<br />
Blue/Cascade blue/Alexa405, and AmCyan/Pacifi c Orange/<br />
Alexa430. <strong>The</strong> 60 mW 355 nm solid-state UV laser has two<br />
fl uorescence detectors (in a<br />
trigon confi guration),<br />
which can detect Hoechst,<br />
Indo-1 (Ca-bound and<br />
unbound) for Ca2+-fl ux<br />
measurements, and DAPI.<br />
This instrument is<br />
also equipped with a<br />
programmable HTS (high<br />
throughput screening<br />
device) for automated<br />
collection <strong>of</strong> data from 96-<br />
384 well microplates.<br />
Flow Cytometer:<br />
FACSCalibur (Sort)<br />
(BD Biosciences)<br />
<strong>The</strong> FACSCalibur fl ow<br />
cytometer is equipped with<br />
two lasers, a 488 nm blue<br />
and 635 nm red, and four<br />
fl uorescence detectors in<br />
addition to forward and side scatter. <strong>The</strong> instrument will be<br />
located in the recently completed Cancer Biology Building. This<br />
instrument has limited sorting capability but because <strong>of</strong> the<br />
relatively slow sort speed is used primarily for analysis.<br />
For additional information contact:<br />
Dan Rosson, Ph.D.<br />
Flow Cytometry<br />
Molecular <strong>Science</strong>s Building, Rm. 214<br />
858 Madison Ave.<br />
(901) 448-4279<br />
drosson@utmem.edu<br />
5
6<br />
Genomics and Bioinformatics Cores<br />
Transgenic/Knockout Core<br />
<strong>The</strong> transgenic/knockout facility at <strong>The</strong> <strong>University</strong> <strong>of</strong><br />
<strong>Tennessee</strong>, <strong>Health</strong> <strong>Science</strong> <strong>Center</strong>, is equipped with all the<br />
necessary hoods, incubators, microscopes, micro injectors<br />
and micromanipulators to produce embryonic stem cells and<br />
transgenic and knockout mice. <strong>The</strong> facility is in operation and<br />
producing KO and transgenic mice and also provides advice<br />
and expertise to interested faculty. <strong>The</strong> facility currently has<br />
one Ph.D.-level, full-time staff person that generates transgenic<br />
and KO/chimeric mice for the <strong>University</strong> <strong>of</strong> <strong>Tennessee</strong> <strong>Health</strong><br />
<strong>Science</strong> <strong>Center</strong>.<br />
For additional information contact:<br />
Ioannis Dragatsis, Ph.D., Director<br />
Physiology<br />
idragats@utmem.edu<br />
(901) 448-3615 � (901) 448-5822 (alternate)<br />
<strong>The</strong> DNA Discovery Core<br />
This Core has three elements:<br />
� Service: <strong>The</strong> core will provide genome screening and<br />
polymorphic detection for research and education in UT<br />
and other institutions in <strong>Tennessee</strong>. Meanwhile, the core<br />
will continue develop and modify protocols for faster<br />
processes and lower costs during services. <strong>The</strong> <strong>Center</strong> has<br />
purchased a SpectruMedix genetic analysis machine to aid<br />
in the detection <strong>of</strong> genetic abnormalities.<br />
� Research: <strong>The</strong> core serves as a resource for the<br />
development <strong>of</strong> research projects that involves in genetic<br />
mapping, fi ne mapping, genome screening, DNA<br />
sequencing, and positional cloning.<br />
� Education: <strong>The</strong> core will serve as an education and<br />
training base for genetic and genomic analyses. Technique<br />
training currently includes genomic analysis and genome<br />
comparison, genome screening, simple sequence repeat<br />
length polymorphism (SSLP) analysis, Single nucleotide<br />
polymorphism (SNP) detection, and DNA sequencing.<br />
For additional information contact:<br />
Weikuan Gu, Ph.D., Director<br />
Orthopaedic Surgery<br />
wgu@utmem.edu<br />
(901) 448-2259 � (901) 448-5880 (alternate)<br />
Mouse Resource Core for Cancer Genetics<br />
<strong>The</strong> aims <strong>of</strong> this Core include:<br />
� Maintain and distribute to our researchers a wide variety<br />
<strong>of</strong> common strains <strong>of</strong> mice that have been critical in<br />
cancer biology.<br />
� Generate novel mouse strains as part <strong>of</strong> the Complex<br />
Trait Consortium. <strong>The</strong>se lines will be distributed to the<br />
cancer biologist.<br />
� Generate new mutants in key genes known to be involved<br />
in cancer progression, surveillance, metastasis using novel<br />
methods developed at ORNL and UTHSC.<br />
� Core facilities for genotype verifi cation and quality<br />
assurance.<br />
� Core mouse bioinformatics module to allow investigators<br />
simple tools to track mouse breeding and use in<br />
experimental protocols.<br />
For additional information contact:<br />
Robert Williams, Ph.D., Director<br />
Anatomy and Neurobiology<br />
rwilliam@nb.utmem.edu<br />
(901) 448-7050 � (901) 448-5965 (alternate)<br />
Bioinformatics Core<br />
In addition to Dr. Cui, the Core’s faculty includes Julia Krushkal<br />
Ph.D. and Rongling Li, Ph.D. who have expertise in statistical<br />
genetics and genetic epidemiology. <strong>The</strong> Core is in the process <strong>of</strong><br />
buying a Linux Beowulf cluster with at least 32 Intel Pentium<br />
Xeon 2.4GMHz processors. This will provide the computational<br />
power to explore the core problems <strong>of</strong> Bioinformatics and<br />
Computational Biology like genetic network modeling; protein<br />
structure prediction and genome-wide DNA sequence analysis.<br />
Researchers will have access to expert help in the analysis <strong>of</strong><br />
regulatory networks from genome-wide gene expression data.<br />
A programmer and a systems administrator staff the Core.<br />
Drs. Krushkal and Li have developed and are teaching<br />
introductory and advanced courses in Bioinformatics. <strong>The</strong>se<br />
would be open to <strong>Center</strong> personnel.<br />
For additional information contact:<br />
Yan Cui, Ph.D., Director<br />
Molecular <strong>Science</strong>s<br />
ycui2@utmem.edu<br />
(901) 448-3240 � (901) 448-6150 (alternate)
Proteomics Core<br />
This core helps researchers identify protein expression patterns<br />
that are altered in development, growth, aging and disease.<br />
<strong>The</strong> <strong>Center</strong>, in conjunction with the College <strong>of</strong> Medicine, has<br />
invested in two state <strong>of</strong> the art mass spectrometers to assist in<br />
the identifi cation <strong>of</strong> protein expression, a workstation for the<br />
visualization <strong>of</strong> protein spots, s<strong>of</strong>tware for the determination<br />
<strong>of</strong> protein differences, and a robotics system for the excision<br />
and processing <strong>of</strong> proteins for mass spectroscopic analysis.<br />
Currently, the Laboratory for Protein Analysis and Proteomics<br />
is equipped with the following mass spectrometers: a Bruker<br />
Ultrafl ex matrix-assisted-laser-desorption/ionization-<br />
Time-<strong>of</strong>-Flight refl ector mass spectrometer (MALDI-ToFToF)<br />
and a Sciex API (Atmospheric Pressure Ionization) QStar DE<br />
electrospray mass spectrometer. <strong>The</strong> MALDI-ToFToF is the<br />
primary instrument for proteomics identifi cations from<br />
polyacrylamide.<br />
T<br />
he Imtek MicroCAT II scanner is an X-ray computed<br />
tomography system capable <strong>of</strong> performing<br />
non-invasive imaging studies <strong>of</strong> tissue specimens or<br />
rats or mice in vivo. Sequential scans over time can<br />
be done using the same animals<br />
thereby reducing the number<br />
needed per experiment. <strong>The</strong><br />
imaging system with a 60 kVp<br />
x-ray source and 1024 x<br />
1024-pixel CCD detector<br />
provides a 50 mm diameter x<br />
50 mm long cylindrical fi eld<br />
<strong>of</strong> view (mouse), or 100 mm<br />
diameter x 50 mm long FOV<br />
for rat studies, with 50 micron<br />
(10% MTF) reconstructed<br />
image resolution. This<br />
translates into a six-minute<br />
scan <strong>of</strong> a whole mouse. <strong>The</strong><br />
source can also be operated at<br />
a maximum voltage <strong>of</strong> 90kVp<br />
to provide a 10-micron focal<br />
spot. With this option, the<br />
scanner can be re-confi gured<br />
by the user to provide 6-10micron<br />
resolution (10% MTF)<br />
reconstructed images. Gantry includes a four-axis precision<br />
motion control system, vibration isolation, a fully interlocked<br />
FDA compliant x-ray cabinet, and a removable mouse/<br />
specimen bed. <strong>The</strong> system computer is a dual processor<br />
<strong>The</strong> Proteomics laboratory <strong>of</strong>fers customized support and<br />
advice for every individual investigator desiring access. <strong>The</strong><br />
format <strong>of</strong> the laboratory permits users to quickly become<br />
advanced and heavy users <strong>of</strong> proteomics in their research.<br />
This didactic and practical experience gained by investigators<br />
in the Proteomics laboratory ensures that protein mass<br />
spectrometry will be used effectively and accurately as a<br />
powerful research tool in the College <strong>of</strong> Medicine at the<br />
<strong>University</strong> <strong>of</strong> <strong>Tennessee</strong>, Memphis.<br />
For additional information contact:<br />
George Hilliard, Ph.D., Director<br />
Molecular <strong>Science</strong>s<br />
ghilliard@utmem.edu<br />
(901) 448-6779 � (901) 448-6150 (alternate)<br />
www.utmem.edu/proteomics<br />
Imtek MicroCat/SPECT System<br />
All microCAT systems can be confi gured with optional high<br />
resolution SPECT detector heads for multi-modality studies.<br />
With the optional variable focus x-ray source, images down to 15<br />
micron resolution can be acquired in vivo.<br />
2 GHz Pentium IV workstation with 1 GB RAM, >100 GB<br />
RAID storage and an ultra-high resolution fl at-panel display.<br />
Data acquisition, image reconstruction, and Amira TM<br />
(Indeed-S<strong>of</strong>tware GmbH) Advanced 3D Visualization and<br />
Volume Modeling s<strong>of</strong>tware<br />
packages are provided.<br />
System operation and scanning<br />
<strong>of</strong> specimens/animals is provided<br />
by trained personnel or user and<br />
s<strong>of</strong>tware analyses training can be<br />
arranged by appointment.<br />
<strong>The</strong> MicroCAT system<br />
confi guration includes a dualhead<br />
single photon emission<br />
tomography (SPECT) data<br />
acquisition system so monitoring<br />
and distribution <strong>of</strong> radiolabeled<br />
compounds can be superimposed<br />
on MicroCT scans. Micro-SPECT<br />
will enable three-dimensional<br />
cross-sectional imaging <strong>of</strong><br />
distribution <strong>of</strong> radiolabel in<br />
small laboratory animals or tissue<br />
specimens at a spatial resolution<br />
in the micron \range. This dual<br />
MicroCAT+ SPECT functionality provides high resolution,<br />
high sensitivity, in vivo imaging to detect and quantify<br />
anatomical and functional changes associated with disease<br />
progression, and evaluate therapeutic effi cacy.<br />
7
8<br />
Key Features:<br />
� Superior image quality due<br />
to low-noise CCD detectors<br />
� Unique capability to adjust<br />
x-ray source and detector<br />
positions for high resolution and<br />
Field <strong>of</strong> View (FOV)<br />
� Multiple options for high<br />
speed 3-D reconstruction<br />
� Optional high resolution SPECT<br />
detector for integrated SPECT/CT<br />
Representative Studies:<br />
� Skeletal biology and disease:<br />
provides accurate calibrated<br />
measures <strong>of</strong> bone mineral density<br />
(BMD) and other structural<br />
parameters, 3-D rendering <strong>of</strong><br />
trabecular architecture and bone/<br />
biomaterial interfaces.<br />
� Joint degeneration: characterizes<br />
anatomical changes in joint<br />
space, and density/architectural<br />
changes in subchondral bone,<br />
measurement <strong>of</strong> s<strong>of</strong>t tissue edema.<br />
� Oncology: measures solid tumor<br />
volume and quantify metastases;<br />
<strong>University</strong> <strong>of</strong> Memphis<br />
Integrated Microscopy <strong>Center</strong><br />
T<br />
he Integrated<br />
Microscopy <strong>Center</strong> is a<br />
fee-for-service facility<br />
on the <strong>University</strong> <strong>of</strong><br />
Memphis main campus since 1976.<br />
<strong>The</strong> facility houses a variety <strong>of</strong> light<br />
and electron microscopes and a<br />
highly trained staff.<br />
Available equipment:<br />
� FEI XL30 Environmental<br />
Scanning Electron Microscope<br />
with an EDAX energy dispersive<br />
X-ray system<br />
� JEOL JEM1200EX II<br />
transmission electron<br />
microscope with an AMT 2K<br />
differentiates normal tissues from<br />
tumors through analysis <strong>of</strong> vascularity.<br />
� Cardiovascular and respiratory<br />
disease: characterizes anatomical<br />
differences in tissue, organ, such as<br />
airway structures, vessel geometry, etc.<br />
� Functional imaging with integrated<br />
microCT/SPECT: provides insight into<br />
biologic processes in living animals by<br />
imaging the deposition <strong>of</strong> labeled<br />
antibodies or uptake <strong>of</strong> labeled<br />
molecules. <strong>The</strong> disease progression as<br />
well as therapeutic intervention can be<br />
studied in a single animal.<br />
For more information or to<br />
arrange an appointment,<br />
contact:<br />
Jinsong Huang, Ph.D.<br />
Orthopaedic Surgery<br />
(901) 523-8990 ext 6444 (Laboratory)<br />
(901) 647-7909 (Mobile)<br />
jhuang4@utmem.edu<br />
Bottom Mount CCD Camera<br />
� Nikon CI Confocal Laser<br />
Scanning microscope with three<br />
laser lines, refl ectance and a<br />
transmitted detector<br />
� Nikon E800 research microscope<br />
with a Nikon 12 million pixel<br />
camera<br />
� Digital Instrument NanoScope<br />
IIIa scanning probe microscope<br />
and a Digital Instrument Bioscope<br />
for cell imaging<br />
� Balzers BAF 301 Freeze Fracture<br />
Unit<br />
� Balzers FSU Freeze Substitution<br />
Unit
� Balzers HMP 010 High Pressure Freezer<br />
� Equipment for thinning metal for TEM<br />
Our technical staff is skilled in the operation <strong>of</strong> all <strong>of</strong> our<br />
equipment and in preparing the samples for you. We have made<br />
it a policy in the IM <strong>Center</strong> to be fl exible about client use <strong>of</strong> our<br />
equipment and are willing to train clients at their request.<br />
We have a complete TEM prep lab, which has several ultra<br />
microtomes, including one with cryo capabilities and a Lynx<br />
tissue processor. We can handle both research and clinical<br />
electron microscopy and are pr<strong>of</strong>i cient in negative staining<br />
techniques. Our research histology lab is set up to handle a<br />
variety <strong>of</strong> tissue types and give special handling for diffi cult<br />
specimens. We do both frozen and paraffi n samples. We also<br />
have protocols for immunohistochemistry.<br />
<strong>The</strong> <strong>University</strong> <strong>of</strong> Memphis also has other facilities on campus<br />
that <strong>of</strong>fer services. Equipment available on a fee-for-service<br />
basis from other departments.<br />
For additional information contact:<br />
Lou Boykins<br />
101 Life <strong>Science</strong> Bldg<br />
3774 Walker Ave<br />
Memphis TN 38152<br />
(901) 678-2034<br />
lboykins@memphis.edu<br />
http://imc.memphis.edu/<br />
Biomedical Engineering<br />
Eugene Eckstein<br />
(901) 678-3505 �Eckstein@memphis.edu<br />
� Triboscope nanoindenter – Measures hardness<br />
Ernö Lindner<br />
(901) 678-5641 � elindner@memphis.edu<br />
� EcoChemie Surface Plasmon Resonance ET ___ for<br />
protein adsorption<br />
� Asylum Research MFP-3D Atomic Force Microscope for<br />
high resolution <strong>of</strong> fl at surface<br />
Warren Haggard<br />
(901) 678-4346 � whaggrd1@memphis.edu<br />
� Abbott TDX (ET 328) – Antibiotic measurements<br />
(selected)<br />
Warren Haggard/ Joel Bumgardner<br />
� Varian Gel Permeation Chromatography ET 322 – MW<br />
measurements <strong>of</strong> polymers<br />
� Differential Scanning Calorimetry ET 316 – Tg & thermal<br />
properties<br />
� Instron Tensile Testing Machine ET 301A – Mech.<br />
Properties (900 lb max load) 9
10<br />
Physics Department<br />
Sanjay Mishra<br />
(901) 678-3115<br />
srmishra@memphis.edu<br />
� Bruker D8 Advance X-ray Diffractometer* – Analyze phase<br />
composition and structure <strong>of</strong> materials<br />
� DuPont 910 DSC** – Phase<br />
transition study <strong>of</strong> materials<br />
M. Shah Jahan<br />
(901) 678-2620<br />
mjahan@memphis.edu<br />
� Varian E4 ESR/EPR<br />
Spectrometer – Free radicals<br />
analyzer<br />
� Bruker EMX 300 ESR/EPR<br />
Spectrometer*** – Free<br />
radical analyzer<br />
� Varian Cary 5E UV-Vis-NIR<br />
Spectrophotometer –<br />
optical density, absorption,<br />
transmission, refl ection<br />
� <strong>The</strong>rmoluminescence<br />
(Harsaw-Bicron) – defects<br />
in solids, chemiluminescence,<br />
thermal activation<br />
energy, radiation dosimetry<br />
*<strong>The</strong> Bruker D8 Advance X-ray<br />
Diffractometer is the state <strong>of</strong> the art<br />
equipment to analyze phase,<br />
composition, and structure <strong>of</strong><br />
materials. <strong>The</strong> system is capable <strong>of</strong><br />
doing fast measurements on variety<br />
<strong>of</strong> materials such as metals, ceramic,<br />
organic materials, and thin fi lms.<br />
<strong>The</strong> system is equipped with a<br />
high temperature stage (Room<br />
temperature-1400C) and has<br />
powerful analysis s<strong>of</strong>tware.<br />
**DuPont 910 DSC: This Differential Scanning Calorimeter system is<br />
capable <strong>of</strong> performing phase transition study <strong>of</strong> materials especially<br />
polymers and metals. It has a dynamic temperature range <strong>of</strong> RT-600C.<br />
***Bruker EMX system is fully automated, high-sensitive, state-<strong>of</strong>-the-art<br />
free-radical analyzer<br />
W. Harry Feinstone <strong>Center</strong> for Genomic Research<br />
<strong>The</strong> W. Harry Feinstone <strong>Center</strong> for Genomic Research provides<br />
equipment and support for all Affymetrix products using the<br />
Scanner 3000 with 7G upgrade. Our technical staff is skilled<br />
in the operation <strong>of</strong> this equipment and in data management<br />
and analysis. Through consultation and collaboration we can<br />
provide you with methods development, study design, data<br />
acquisition and analysis, including production <strong>of</strong> fi gures for<br />
publication. GeneChip applications include RNA expression,<br />
splice variant detection, and Chip-on-Chip assays. In addition,<br />
100k and 500k SNP chips support analysis <strong>of</strong> chromosome<br />
structure and loss <strong>of</strong> heterozygosity, as well as, genetic<br />
association studies.<br />
<strong>University</strong> <strong>of</strong> Memphis<br />
Bionformatics<br />
<strong>The</strong> Bioinformatics Program<br />
at the <strong>University</strong> <strong>of</strong> Memphis<br />
(http://cas.memphis.edu/binf/)<br />
is composed <strong>of</strong> over a dozen<br />
faculty members with a wide<br />
range <strong>of</strong> research interests.<br />
<strong>The</strong> Program consists <strong>of</strong> four<br />
service units:<br />
� Genomic and proteomic<br />
design and analysis<br />
� Database design and<br />
implementation<br />
� Gene function and pathway<br />
analysis<br />
� Meta-analysis and<br />
integrative statistics<br />
Each unit is headed by a faculty<br />
member who supervises<br />
graduate students and staff who<br />
are available to collaborate with<br />
researchers in the community.<br />
<strong>The</strong> Bioinformatics <strong>Center</strong> has<br />
access to the High Performance<br />
Computing facility at the<br />
FedEx Institute <strong>of</strong> Technology<br />
(http://itd.memphis.edu/<br />
researchcomputing.htm). In<br />
addition, the <strong>Center</strong> supports<br />
many commercially available statistical and analytical s<strong>of</strong>tware<br />
packages as well as several in-house databases and<br />
bioinformatic tools.<br />
For more information contact:<br />
Thomas R. Sutter, Ph.D.<br />
3774 Walker Ave<br />
Memphis TN 38152<br />
(901) 678-8391<br />
tsutter@memphis.edu<br />
For parking information, visit www.memphis.edu and search for<br />
the Zach Curlin parking garage.
L<br />
aser Capture Microdissection (LCM) provides a<br />
rapid, reliable, one-step method to obtain pure<br />
populations <strong>of</strong> targeted cells from specifi c<br />
microscopic regions for subsequent analysis.<br />
LCM was developed to enable<br />
investigators and clinicians to perform<br />
tissue microdissection on a routine<br />
basis. <strong>The</strong>re are no limitations in the<br />
ability to amplify DNA or RNA from<br />
individual cells or cell clusters (e.g.,<br />
tumors) from tissue sections, blood<br />
smears, or cell cultures. LCM has been<br />
used to explore gene expression, loss<br />
<strong>of</strong> heterozygosity, micro-satellite<br />
instability, clonality and for<br />
proteomics.<br />
<strong>The</strong> Arcturus Pixcell II LCM at the<br />
UTHSC facility has major advantages<br />
compared to other instruments in that<br />
1) the laser never touches the tissue<br />
itself, thereby avoiding possible heat<br />
shock and damage, 2) cells are<br />
captured directly onto<br />
an optically transparent<br />
cap for reliable transfer<br />
<strong>of</strong> every sample to the a<br />
ppropriate assay tube, and<br />
3) the accuracy <strong>of</strong> each<br />
step <strong>of</strong> cell capture can<br />
be digitally documented<br />
by the accompanying<br />
archiving s<strong>of</strong>tware. <strong>The</strong><br />
PixCell II also has a<br />
fl uorescence system with<br />
fi lter cubes for blue (Ex<br />
455-495 nm/Em >510<br />
nm), green (Ex 503-547<br />
nm/Em >565 nm), and red<br />
(Ex 590-650 nm/Em >667<br />
nm) wavelengths. <strong>The</strong><br />
following fl uorophores<br />
have been shown to be<br />
compatible with LCM:<br />
Cy2, Cy3, Cy5; Alexa 488,<br />
532, 546, 568; FITC; Oregon Green; propidium iodide; TRITC;<br />
phycoerythrin; rhodamine; To-Pro-3; and, Texas Red.<br />
Laser Capture Microdissection<br />
Figure 1. Diagrammatic representation <strong>of</strong> laser<br />
capture microdissection steps. Reprinted from<br />
Arcturus 2002 Web site (www.arctur.com).<br />
Procedures<br />
Cells can only be captured from dehydrated tissue sections or<br />
cell samples centered on a glass slide.<br />
<strong>The</strong> proprietary CapSure, an optically transparent cap<br />
containing a thermoplastic membrane<br />
transfer fi lm, is placed on the slide and<br />
cells are dissected by the focal melting<br />
<strong>of</strong> the membrane through laser<br />
activation (fi g 1). This short, low<br />
power infrared laser pulse, with beam<br />
sizes <strong>of</strong> 7.5, 15 and 30 µm, is triggered<br />
with a push <strong>of</strong> a button on the joystick.<br />
At no time does the laser directly touch<br />
the tissue sample; therefore, neither the<br />
quality <strong>of</strong> nucleic acids and proteins<br />
within the sample nor cell morphology<br />
are compromised, and the surrounding<br />
tissue remains intact on the slide.<br />
<strong>The</strong> captured cells remain attached to<br />
the transfer fi lm surface on the<br />
CapSure cap, which is then placed<br />
directly into a microcentrifuge<br />
tube containing the<br />
user-identifi ed extraction<br />
buffer for specifi c assay(s).<br />
Publication quality,<br />
digitized images can be<br />
taken <strong>of</strong> the tissue section<br />
before and after LCM, as<br />
well as <strong>of</strong> the selected<br />
tissue retained on the<br />
CapSure cap, to document<br />
the specifi city <strong>of</strong> each<br />
sample (fi g. 2).<br />
Extraction <strong>of</strong> DNA has<br />
been demonstrated using<br />
both ethanol fi xed and<br />
formalin fi xed paraffi n<br />
embedded tissue; RNA can<br />
be recovered from fresh<br />
frozen tissue sections.<br />
Guidelines for amounts<br />
needed per sample for molecular analyses are: 10-20 cells from<br />
a 10-micron section has been shown to work for PCR; 50,000<br />
cells for 2-D gel a nalysis; and, 20,000 cells for a western blot.<br />
Figure 2. Digitized images <strong>of</strong> each step <strong>of</strong> laser capture microdissection. (A) Rat<br />
brain was fresh frozen and 7 µm cryosections were briefl y Nissl stained, then<br />
rapidly dehydrated and laser-captured. (B) Paraffi n-embedded rat kidney 10 µm<br />
sections were briefl y stained with H&E, then rapidly dehydrated and microdissected.<br />
<strong>The</strong> last panel for each tissue is an image <strong>of</strong> the captured neurons (A) or<br />
glomerulus (B) adhered to the polymer transfer fi lm on the cap.<br />
11
12<br />
Since slide preparation is the most critical component <strong>of</strong> successful<br />
laser capture, the Director <strong>of</strong> the LCM facility,<br />
Dr. Shannon Matta, will work closely with each investigator to<br />
develop optimal procedure(s) for tissue preparation specifi c to<br />
the sample. In addition, every scientist, physician or pathologist<br />
who wishes to use the LCM system must fi rst be trained and<br />
certifi ed by Dr. Matta.<br />
Mass Spectrometry Core Laboratory<br />
T<br />
he new UTHSC Mass Spectrometry Core<br />
Laboratory will: (a) provide to funded UTHSC<br />
investigators state-<strong>of</strong>-the-art analytical<br />
methodologies; (c) train UTHSC research<br />
personnel; and (d) promote biological mass<br />
spectrometry to the UTHSC research community and other<br />
institutions. <strong>The</strong> center <strong>of</strong> this new UTHSC Core is the Charles<br />
B. Stout Neuroscience Mass Spectrometry Laboratory. <strong>The</strong><br />
laboratory, headed by Dr. Dominic Desiderio, is an established<br />
university, national, and international mass spectrometry center<br />
that has conducted independent and collaborative research,<br />
and has provided mass<br />
spectrometry service to<br />
scientists at UTHSC and<br />
neighboring institutions for<br />
28 years. <strong>The</strong> Associate<br />
Directors are Dr. Giorgianni,<br />
Ph.D., Assistant Pr<strong>of</strong>essor in the<br />
Department <strong>of</strong> Neurology, and<br />
Sarka Beranova-Giorgianni,<br />
Ph.D., Assistant Pr<strong>of</strong>essor <strong>of</strong><br />
Pharmaceutical <strong>Science</strong>s. A<br />
Technologist with expertise in<br />
mass spectrometry will conduct<br />
protein identifi cation/<br />
characterization experiments<br />
that include mass spectrometry<br />
measurements, database searches, and data<br />
interpretation. This MS Core will provide identifi cation <strong>of</strong><br />
proteins from 2D gel spots or from bands from 1D gels via<br />
MS/MS and searches <strong>of</strong> protein sequence databases.<br />
Instrumentation<br />
� Matrix-assisted laser desorption/ionization (MALDI) linear<br />
trap mass spectrometer; MALDI-LTQ (<strong>The</strong>rmoelectron)<br />
� Liquid chromatography-nanoelectrospray linear trap mass<br />
spectrometer; LC-nanoESI-LTQ (<strong>The</strong>rmoelectron)<br />
For additional information, contact:<br />
Shannon Matta, Ph.D., Director<br />
Pharmacology<br />
(901) 448-2874 � (901) 448-6000 (alternate)<br />
smatta@utmem.edu<br />
www.utmem.edu/lcm<br />
LCM and the PixCell II instrument details are available on the<br />
Arcturus Pixcell II page – www.arctur.com/research_portal/ products/<br />
pixcell_main.htm<br />
� Liquid chromatography-nanoelectrospray ion trap mass<br />
spectrometer; LCQDeca (<strong>The</strong>rmoelectron)<br />
� Liquid chromatography-nanoelectrospray quadrupole<br />
time-<strong>of</strong>-fl ight (QTOF) mass spectrometer; Q-TOF2<br />
(Waters)<br />
� Matrix-assisted laser desorption/ionization time-<strong>of</strong>-fl ight<br />
(MALDI TOF) mass spectrometer (Applied Biosystems).<br />
This user-friendly instrument provides the molecular<br />
weight (MW) <strong>of</strong> individual tryptic peptides in an<br />
unseparated protein digest.<br />
Liquid chromatography<br />
Three nanoLC systems are<br />
interfaced with the mass<br />
spectrometers for online LC-<br />
MS/MS analyses: Surveyor<br />
nanoLC (<strong>The</strong>rmoelectron);<br />
FAMOS/SWITCHOS/Ultimate<br />
nanoLC (Dionex); and CapLC<br />
nanoLC (Waters). <strong>The</strong> Dionex<br />
system has multidimensional<br />
liquid chromatography<br />
(MDLC) capabilities.<br />
Bioinformatics<br />
Each mass spectrometer<br />
includes a computer for<br />
instrument control and data acquisition. In addition, there are<br />
four computers dedicated to database searches. Search engines<br />
for in-house database searches include two copies <strong>of</strong> SEQUEST<br />
Browser, and two copies <strong>of</strong> Bioworks 3.2. <strong>The</strong> Core will use<br />
free online bioinformatics resources such as MASCOT (free<br />
version), Expasy, and Scansite.<br />
<strong>The</strong> Core will provide support for currently funded<br />
UTHSC investigators through access to advanced MS<br />
instrumentation and expert assistance. <strong>The</strong> Core will help to
initiate new research projects that will be enabled by<br />
state-<strong>of</strong>-the-art MS technologies and expertise.<br />
<strong>The</strong> Core will train students, postdoctoral fellows, and residents<br />
in mass spectrometry. Drs. Desiderio and Beranova-Giorgianni<br />
teach a course on the Basic Principles <strong>of</strong> Mass Spectrometry<br />
in the Department <strong>of</strong> Molecular <strong>Science</strong>s, organize discussion<br />
groups, and present seminars to promote mass spectrometry<br />
and to attract new users.<br />
Molecular Resource <strong>Center</strong> Services<br />
� DNA Sequencing<br />
� Genetic Analysis/Genotyping<br />
� Phosphor-imaging<br />
� Real Time PCR<br />
� Confocal Microscopy<br />
� Cell Sorting<br />
� Microarray Facility<br />
� Fluorescence Microscopy<br />
� Primer Express S<strong>of</strong>tware<br />
DNA Sequencing<br />
<strong>The</strong> MRC currently has ABI PRISM<br />
310 and 3130xl Genetic Analyzers<br />
for DNA sequencing and fragment<br />
analysis. <strong>The</strong> 3130s have 16 capillaries<br />
that operate in parallel and are fully<br />
automated from sample loading to<br />
data analysis. We currently employ<br />
these two instruments for sequencing<br />
and genotyping. <strong>The</strong> 310 is a single<br />
capillary instrument that can support<br />
most gel-based assays. It is currently<br />
used for fragment analysis employing<br />
fl uorescent markers that precede the<br />
ones used with the 3130xl. As part <strong>of</strong><br />
our DNA sequencing services, we <strong>of</strong>fer<br />
plasmid miniprep purifi cation for<br />
sequencing only. Details on<br />
submission <strong>of</strong> transformed bacteria<br />
and costs can be obtained from MRC<br />
personnel.<br />
<strong>The</strong> MRC has an ABI PRISM 7700 Sequence Detection System<br />
for Real Time PCR. This instrument includes a built in 9600<br />
thermalcycler, a laser to induce fl uorescence and real-time<br />
detection s<strong>of</strong>tware. Some <strong>of</strong> the applications <strong>of</strong> the ABI PRISM<br />
Molecular Resource <strong>Center</strong><br />
ABI PRISM 3130xl Genetic Analyzer<br />
Light Cycler®480; Universal Probe Library<br />
For additional information, contact:<br />
Dominic M. Desiderio, Ph.D., Director<br />
Francesco Giorgianni, Ph.D., Associate Director<br />
Sarka Beranova-Giorgianni, Ph.D., Associate Director<br />
Neurology<br />
(901) 448-6199<br />
7700 include real-time quantitation, SNP detection, and +/-<br />
assays with internal positive control.<br />
<strong>The</strong> ABI Primer Express s<strong>of</strong>tware allows you to independently<br />
design and analyze oligo sequences for a variety <strong>of</strong> PCR and<br />
sequencing applications.<br />
Real Time PCR<br />
<strong>The</strong> LightCycler® 480 (LC480)<br />
Instrument is a rapid, thermal blockcycler<br />
with integrated real-time, online<br />
detection capabilities. Based on a<br />
comprehensive improvement <strong>of</strong> the<br />
Peltier-based block technology, the<br />
Instrument provides extraordinary<br />
well-to-well temperature homogeneity<br />
and maximized inter-well, inter-cycle<br />
reproducibility. <strong>The</strong> special arrangement<br />
<strong>of</strong> optical components in the<br />
LC480 Instrument ensures the uniform<br />
collection <strong>of</strong> signals across the<br />
plate and makes analysis independent<br />
<strong>of</strong> the sample position on the plate.<br />
<strong>The</strong> LightCycler® 480 System setup<br />
enables the use <strong>of</strong> all current probe<br />
formats (e.g., SYBR Green I,<br />
Hydrolysis Probes, HybProbe Probes)<br />
and provides the ideal solution for fast<br />
and precise qualitative or quantitative<br />
detection <strong>of</strong> nucleic acids, genotyping,<br />
and mutation analysis. <strong>The</strong> ability to<br />
freely combine fi ve excitation and six<br />
emission fi lters permits analysis <strong>of</strong> signals from multiple dyes<br />
(e.g., LightCycler® Red 610, 640, LightCycler® CYAN 500, FAM,<br />
VIC, HEX, Cy5, and Fluorescein) in monocolor as well as in<br />
multiplex assays.<br />
13
14<br />
<strong>The</strong> user-friendly LightCycler® 480 S<strong>of</strong>tware enables highly<br />
fl exible and extensive data analysis for different scientifi c needs.<br />
Based on the modular design <strong>of</strong> the s<strong>of</strong>tware, the user<br />
can customize the system by adding<br />
advanced analysis modules (e.g.,<br />
LightCycler® 480 Relative<br />
Quantifi cation S<strong>of</strong>tware, LightCycler®<br />
480 Genotyping S<strong>of</strong>tware) to the<br />
LightCycler® 480 Basic S<strong>of</strong>tware (run<br />
option, Tm calling option, absolute<br />
quantifi cation option).<br />
Phosphor-imaging<br />
<strong>The</strong> Molecular Dynamics Typhoon<br />
Phosphor-imager is capable <strong>of</strong><br />
scanning exposed storage phosphor<br />
screens, fl uorescent (532nm or 633nm<br />
excitation), and chemiluminescent<br />
samples. ImageQuant s<strong>of</strong>tware<br />
analysis tools can be used to<br />
perform area or volume quantitation<br />
on scanned images. DNA and protein<br />
fragments separated by electrophoresis<br />
can be analyzed using the fragment<br />
analysis s<strong>of</strong>tware.<br />
Fluorescence Microscope<br />
<strong>The</strong> Axiophot Photomicroscope is<br />
available for either transmitted light<br />
or fl uorescence detection on stained<br />
slides. Optronics Magnafi re s<strong>of</strong>tware<br />
can be used to capture and print<br />
images, using a digital camera, from<br />
bright fi eld monochrome (grayscale)<br />
images and color images as well as<br />
fl uorescent images.<br />
Microarray Production<br />
Production <strong>of</strong> microarrays is <strong>of</strong>fered<br />
DNA for the arrays can be produced<br />
as either amplifi ed cDNA clones or<br />
oligonucleotides and applied to glass<br />
substrates. Users must supply the appropriate<br />
cDNA clones or sequences for<br />
the oligonucleotides. S<strong>of</strong>tware is<br />
available to aid in the design <strong>of</strong> the appropriate<br />
oligonucleotides. Currently, a mouse 15K array from cDNA<br />
clones supplied by the National Institute <strong>of</strong> Aging is available.<br />
Agilent Bioanalyzer<br />
<strong>The</strong> Bioanalyzer system is an automated, computer controlled,<br />
integrated analyzer capable <strong>of</strong> sizing and concentration<br />
Zeiss Axiophot microscope<br />
Agilent Bioanalyer<br />
Liquid Handling Robot<br />
determinations <strong>of</strong> nucleic acids and proteins. Sample<br />
preparations include PCR and RT-PCR fragments, restriction<br />
digests, total RNA and mRNA, cell lysates, column fractions<br />
and purifi ed proteins.<br />
Liquid Handling Robot<br />
<strong>The</strong> MRC has excess capacity for all<br />
<strong>of</strong> our services. A liquid handling<br />
robot (Perkin Elmer Multiprobe HT<br />
EX) is available to scale up assays<br />
as necessary.<br />
Other MRC Services<br />
To better serve the needs <strong>of</strong> the research<br />
community, the MRC has contracted<br />
services with the following vendors:<br />
� Peptide Synthesis<br />
Biomolecules Midwest<br />
Dr. John Gorka<br />
1-877-939-9676<br />
� Protein Sequencing<br />
Midwest Analytical<br />
Dr. David McCourt<br />
1-800-481-0790<br />
Felicia Waller<br />
(Phosphoimaging & Digital Microscopy)<br />
fwaller@utmem.edu<br />
Molecular Resource <strong>Center</strong><br />
19 S. Manassas 3rd Floor<br />
Memphis, TN 38163<br />
(901) 448-6191 � (901) 448-8229 (fax)<br />
For more information on MRC,<br />
contact:<br />
William Taylor, Ph.D.<br />
Molecular Resource <strong>Center</strong><br />
wtaylor@utmem.edu<br />
Thomas Cunningham, Ph.D.<br />
(DNA Sequencing & Real Time PCR)<br />
tcunningham@utmem.edu
T<br />
he Neuroscience Imaging <strong>Center</strong> within the<br />
Neuroscience Institute/<strong>Center</strong> <strong>of</strong> Excellence,<br />
located on the 3rd fl oor <strong>of</strong> the Link Building at<br />
855 Monroe, <strong>of</strong>fers use <strong>of</strong> equipment and<br />
technical services in the areas <strong>of</strong> transmission electron<br />
microscopy, confocal microscopy, 3-dimensional neuron<br />
tracing and mapping, conventional light microscopy<br />
applications and tissue preparation to all investigators in the<br />
medical and research fi elds in the mid-south area.<br />
<strong>The</strong> JEOL 2000EX transmission electron microscope<br />
normally runs at 60kV, but can run up to 200kV to<br />
examine semi-thin sections. It is equipped with a highresolution<br />
monitor and Hamamatsu Orca HR digital camera,<br />
providing portable, digital images at high resolution, in several<br />
formats, obviating the need for fi lm and darkroom work.<br />
<strong>The</strong> MicrobrightField, Inc. Neurolucida s<strong>of</strong>tware allows<br />
3D reconstruction <strong>of</strong> neurons, serial reconstruction <strong>of</strong><br />
brain sections, and precise cell plotting <strong>of</strong> histological or<br />
vibrotomed serial sections, using a motorized stage with X, Y<br />
and Z encoders.<br />
<strong>The</strong> BioRad MRC 1024 confocal imaging system is equipped<br />
with a Krypton/Argon laser, creating three excitation lines<br />
co-aligned at 488 nm (blue), 568 nm (green), and 647 nm (far<br />
red), enabling the user to effectively employ double or triple<br />
labeling techniques, and with appropriate barrier fi lters, can<br />
minimize bleed-through. Refl ected light and transmitted light<br />
image capture is also available. Image fi les can be saved to CDs<br />
for portability. This system uses the upright Olympus BX50<br />
with objective lenses ranging from 2x to 100x, and a 20x water<br />
immersion lens.<br />
Technical assistance is available with over 35 years experience in<br />
light and transmission electron microscopy. This lab can<br />
process many types <strong>of</strong> tissue, tissue slices, tissue cultures, and<br />
pellets, embedding in Spurr’s resin, Epon, or Epon/Araldite<br />
blends, as well as LRWhite, utilizing a variety <strong>of</strong> embedment<br />
methods, such as Beem or gelatin capsules, fl at molds, and<br />
fl at embedment <strong>of</strong> tissue slices. Negative staining can be<br />
done as well.<br />
For information concerning scheduling and prices,<br />
contact:<br />
Kathy Troughton<br />
(901) 448-5976<br />
www.utmem.edu/neuroscience/imaging-center<br />
Neuroscience Imaging<br />
Immunogold localization <strong>of</strong> neurophysins in dense core granules <strong>of</strong><br />
vasopressin and oxytocin nerve terminals.<br />
Visualizing the spatial overlap <strong>of</strong> developing pathways in<br />
embryonic mouse brain using fl uorescent dyes and confocal laser<br />
scanning microscopy.<br />
15
16<br />
Public Parking Locations at the <strong>University</strong> <strong>of</strong> <strong>Tennessee</strong><br />
<strong>The</strong>re are fi ve public parking lots on the UTHSC campus:<br />
outdoor lots O and V and indoor garages E, H and P.<br />
Directions<br />
O lot is the parking lot located on the south side <strong>of</strong> the Dunn<br />
Dental Building at 875 Union Ave. <strong>The</strong> parking lot is<br />
accessible from Union Avenue, X lot and T lot. In order to gain<br />
entrance to the parking lot, drivers must enter the gated lot by<br />
pulling a ticket from the parking booth that is monitored by a<br />
parking attendant from the hours <strong>of</strong> 9 a.m. until 5 p.m.<br />
V lot is a metered parking location at the corner <strong>of</strong> Madison<br />
and Manassas with a physical address <strong>of</strong> 780 Madison Ave. <strong>The</strong><br />
entrance to V lot is accessible from Manassas Street.<br />
E garage is located on the north side <strong>of</strong> the General Education<br />
Building, which is on the southeast corner <strong>of</strong> Dunlap and<br />
Madison. <strong>The</strong> physical address <strong>of</strong> E garage is 869 Madison Ave.<br />
<strong>The</strong> entrance to the garage is accessible from Madison Avenue.<br />
H garage is located on the east side <strong>of</strong> the Plaza Complex at<br />
930 Madison Ave. It is adjacent to the building. <strong>The</strong> garage has<br />
entrances from both Madison Avenue and Court Street.<br />
P garage is the parking garage adjacent to the Doctor’s Building<br />
located at 66 North Pauline. <strong>The</strong> entrance to the garage is<br />
accessible from Pauline Street.<br />
Metered Parking<br />
Public spaces adjacent to parking meters are located on:<br />
� the west side <strong>of</strong> the Student Alumni <strong>Center</strong>,<br />
� Manassas between Madison and Jefferson,<br />
� Dunlap across the street from the Hyman Administration<br />
Building.<br />
Map Buildings Legend<br />
4 740 Court Building D5<br />
34 910 Madison Building J6<br />
31 920 Madison Building J5<br />
35 930 Madison Building J6<br />
16 Alexander Bulding H6<br />
11 Beale Building G9<br />
3 Boling <strong>Center</strong> for C4<br />
Developmental Disabilities<br />
6 Cancer Research Bldg. D7<br />
30 Coleman College <strong>of</strong><br />
Medicine Bldg. J4<br />
24 Crowe Research Bldg. H8<br />
29 Docs Field Pavilion I11<br />
37 Doctors Offi ce Bldg L4<br />
17 Feurt Pharmacy G7<br />
Research Bldg<br />
15 GEB Garage H6<br />
14 General Education Bldg G6<br />
12 Goodman Family G11<br />
Residence Hall<br />
2 Hyde Engineering B4<br />
23 Hyman Administration G8<br />
18 Johnson Building G7<br />
19 Link Bulding H7<br />
32 Madison Garage K5<br />
33 Madison Place Building J5<br />
13 Molecular <strong>Science</strong>s Bldg G6<br />
21 Mooney Building H8<br />
22 Nash Addition I8<br />
25 Nash Research Bldg. I8<br />
36 Pauline Annex L4<br />
38 Pauline Garage L4<br />
1 Phi Chi Fraternity House B4<br />
28 Physical Plant Shops I10<br />
27 Purchasing and Physical I10<br />
Plant Building<br />
9 Randolph Hall E6<br />
7 Student <strong>Center</strong> Garage E5<br />
8 Student-Alumni <strong>Center</strong> F5<br />
10 Van Vleet Cancer <strong>Center</strong> F5<br />
5 Just Variety Building D5<br />
20 Wittenborg Anatomy H7<br />
1<br />
2<br />
3<br />
4<br />
5<br />
6<br />
7<br />
8<br />
9<br />
10<br />
11<br />
A B C D E F<br />
0 FEET<br />
0<br />
Union Avenue<br />
Jefferson Avenue<br />
MILES<br />
1<br />
2<br />
Madison Avenue<br />
Marshall Street<br />
500<br />
0.<br />
1<br />
Orleans Street<br />
Washington Ave.<br />
Adams Avenue<br />
3<br />
Monroe Avenue Extended<br />
Court Avenue<br />
© 2007 UTCSL<br />
This map may not be reproduced, copied, altered, or distributed by electronic or traditional means<br />
without permission <strong>of</strong> <strong>The</strong> <strong>University</strong> <strong>of</strong> <strong>Tennessee</strong> Cartographic Services Laboratory.<br />
Custom versions can be generated by calling (865) 974-5348.<br />
Cartography by Will Fontanez, Director<br />
Forrest Park<br />
A B C D E F<br />
4<br />
<br />
5<br />
6<br />
Memphis an<br />
Shelby Coun<br />
<strong>Health</strong> Depart<br />
7<br />
Jefferson Avenu<br />
9<br />
Speech<br />
Bldg.<br />
8<br />
Union Avenue<br />
Walnut Street<br />
Manassas Street
<strong>Health</strong> <strong>Science</strong> <strong>Center</strong><br />
d<br />
ty<br />
ment<br />
e<br />
Dunlap Street<br />
10 10<br />
11 11<br />
Beale Street<br />
Dunlap Street<br />
12 12<br />
G H<br />
Mental <strong>Health</strong><br />
Institute, Memphis<br />
Washington Avenue<br />
LeBonheur Children’s<br />
Medical <strong>Center</strong><br />
14 14<br />
17 17<br />
18 18<br />
23 23<br />
13 13<br />
Linden Avenue<br />
15 15<br />
Monroe Avenue<br />
19 19 20 20<br />
26 26<br />
G H<br />
Madison Avenue<br />
16 16<br />
21 21<br />
24 24<br />
Adams Avenue<br />
Regional Medical <strong>Center</strong><br />
at Memphis<br />
Dunlap Street<br />
Hospital Street<br />
22 22<br />
25 25<br />
Beale Street<br />
29 29<br />
I J<br />
State<br />
Mental<br />
<strong>Health</strong><br />
Building<br />
Dobbs<br />
Research<br />
Institute<br />
28 28<br />
27 27<br />
31 31<br />
I J<br />
30 30<br />
33 33<br />
34 34 35 35<br />
Memphis<br />
Bioworks<br />
Foundation<br />
Memphis<br />
Bioworks<br />
Foundation<br />
East Street<br />
Poplar Avenue<br />
Mid-South<br />
Hospital<br />
Dudley Street<br />
32 32<br />
K L M N O P<br />
Regional<br />
Medical <strong>Center</strong><br />
Parking<br />
Memphis<br />
Bioworks<br />
Foundation<br />
Memphis<br />
Bioworks<br />
Foundation<br />
Dudley Street<br />
Pauline Street<br />
Union Avenue<br />
Veterans Administration<br />
Medical <strong>Center</strong><br />
36 36<br />
37 37<br />
BMH<br />
Pauline Street<br />
38 38<br />
Baptist<br />
Memorial<br />
Hospital<br />
Eastmoreland Avenue<br />
Court Avenue<br />
Camila Street<br />
Madison Avenue<br />
Camila Street<br />
Baptist<br />
Memorial<br />
Hospital<br />
Jefferson Avenue<br />
Baptist<br />
Memorial<br />
Hospital<br />
Linden Avenue<br />
K L M N O P<br />
Somerville Street<br />
Waldran Boulevard<br />
1<br />
2<br />
3<br />
4<br />
5<br />
6<br />
7<br />
8<br />
9<br />
10<br />
11<br />
17
Mission<br />
UT <strong>Health</strong> <strong>Science</strong> <strong>Center</strong> aims to improve human health through education, research, clinical care and public service.<br />
<strong>The</strong> UTHSC campuses include colleges <strong>of</strong> Allied <strong>Health</strong> <strong>Science</strong>s, Dentistry, Graduate <strong>Health</strong> <strong>Science</strong>s,<br />
Medicine, Nursing and Pharmacy. Patient care, pr<strong>of</strong>essional education and research are carried out at hospitals<br />
and other clinical sites across <strong>Tennessee</strong>. Endowed pr<strong>of</strong>essorships, Research <strong>Center</strong>s <strong>of</strong> Excellence,<br />
and continuing relationships with research and healthcare facilities across <strong>Tennessee</strong><br />
ensure that both basic science and applied research stay focused on contemporary health topics.<br />
<strong>University</strong> <strong>of</strong> <strong>Tennessee</strong> <strong>Health</strong> <strong>Science</strong> <strong>Center</strong> Administration<br />
Chancellor (Interim)<br />
Hershel P. Wall, M.D.<br />
Chief <strong>of</strong> Staff and Executive Vice Chancellor (Interim)<br />
Kennard D. Brown, M.P.A., J.D.<br />
Deans:<br />
College <strong>of</strong> Allied <strong>Health</strong> <strong>Science</strong>s (Interim)<br />
William R. Frey, Ph.D. Vice Chancellors:<br />
College <strong>of</strong> Dentistry Academic, Faculty and Student Affairs<br />
Russell O. Gilpatrick, D.D.S. Cheryl R. Scheid, Ph.D.<br />
College <strong>of</strong> Graduate <strong>Health</strong> <strong>Science</strong>s Community Affairs (Interim)<br />
Richard D. Peppler, Ph.D. Kennard D. Brown, M.P.A., J.D.<br />
College <strong>of</strong> Medicine Development and Alumni Affairs<br />
Executive Dean and Memphis Dean (Interim) Linda Garceau-Luis , M.B.A., M.A.<br />
Steve J. Schwab, M.D. Finance and Operations<br />
Knoxville Dean Anthony A. Ferrara, C.P.A., M.A.S.<br />
James J. Neutens, Ph.D. <strong>Health</strong> System Affairs<br />
Chattanooga Dean Michael R. Caudle, M.D.<br />
David C. Seaberg, M.D. Research<br />
College <strong>of</strong> Nursing Leonard R. Johnson, Ph.D.<br />
Donna Hathaway, Ph.D.<br />
College <strong>of</strong> Pharmacy Director, Communications and Marketing<br />
Dick R. Gourley, Pharm.D. Sheila Champlin, M.A.