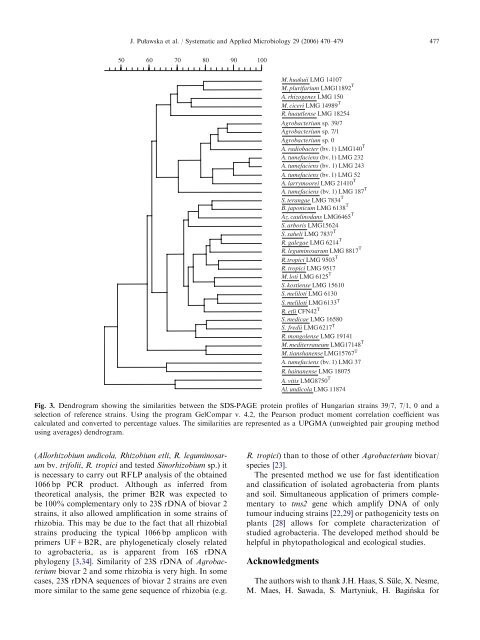
ARTICLE IN PRESSJ. Pu"awska et al. / Systematic <strong>and</strong> Applied Microbiology 29 (2006) 470–479 4775060708090100M. huakuii LMG 14107M. plurifarium LMG11892 TA. rhizogenes LMG 150M. ciceri LMG 14989 TR. huautlense LMG 18254<strong>Agrobacterium</strong> sp. 39/7<strong>Agrobacterium</strong> sp. 7/1<strong>Agrobacterium</strong> sp. 0A. radiobacter (bv. 1) LMG140 TA. tumefaciens (bv. 1) LMG 232A. tumefaciens (bv. 1) LMG 243A. tumefaciens (bv. 1) LMG 52A. larrymoorei LMG 21410 TA. tumefaciens (bv. 1) LMG 187 TS. terangae LMG 7834 TB. japonicum LMG 6138 TAz. caulinodans LMG6465 TS. arboris LMG15624S. saheli LMG 7837 TR. galegae LMG 6214 TR. leguminosarum LMG 8817 TR. tropici LMG 9503 TR. tropici LMG 9517M. loti LMG 6125 TS. kostiense LMG 15610S. meliloti LMG 6130S. meliloti LMG 6133 TR. etli CFN42 TS. medicae LMG 16580S. fredii LMG 6217 TR. mongolense LMG 19141M. mediterraneum LMG17148 TM. tianshanense LMG15767 TA. tumefaciens (bv. 1) LMG 37R. hainanense LMG 18075A. vitis LMG8750 TAl. undicola LMG 11874Fig. 3. Dendrogram showing the similarities between the SDS-PAGE protein pr<strong>of</strong>iles <strong>of</strong> Hungarian strains 39/7, 7/1, 0 <strong>and</strong> aselection <strong>of</strong> reference strains. Using the program GelCompar v. 4.2, the Pearson product moment correlation coefficient wascalculated <strong>and</strong> converted to percentage values. The similarities are represented as a UPGMA (unweighted pair grouping methodusing averages) dendrogram.(Allorhizobium undicola, Rhizobium etli, R. leguminosarumbv. trifolii, R. tropici <strong>and</strong> tested Sinorhizobium sp.) itis necessary to carry out RFLP analysis <strong>of</strong> the obtained1066 bp PCR product. Although as inferred fromtheoretical analysis, the primer B2R was expected tobe 100% complementary only to 23S rDNA <strong>of</strong> biovar 2strains, it also allowed amplification in some strains <strong>of</strong>rhizobia. This may be due to the fact that all rhizobialstrains producing the typical 1066 bp amplicon withprimers UF+B2R, are phylogeneticaly closely relatedto agrobacteria, as is apparent from 16S rDNAphylogeny [3,34]. Similarity <strong>of</strong> 23S rDNA <strong>of</strong> <strong>Agrobacterium</strong>biovar 2 <strong>and</strong> some rhizobia is very high. In somecases, 23S rDNA sequences <strong>of</strong> biovar 2 strains are evenmore similar to the same gene sequence <strong>of</strong> rhizobia (e.g.R. tropici) than to those <strong>of</strong> other <strong>Agrobacterium</strong> biovar/species [23].The presented method we use for fast <strong>identification</strong><strong>and</strong> classification <strong>of</strong> isolated agrobacteria from plants<strong>and</strong> soil. Simultaneous application <strong>of</strong> primers complementaryto tms2 gene which amplify DNA <strong>of</strong> onlytumour inducing strains [22,29] or pathogenicity tests onplants [28] allows for complete characterization <strong>of</strong>studied agrobacteria. The developed method should behelpful in phytopathological <strong>and</strong> ecological studies.AcknowledgmentsThe authors wish to thank J.H. Haas, S. Su¨le,X.Nesme,M. Maes, H. Sawada, S. Martyniuk, H. Bagin´ska for
478ARTICLE IN PRESSJ. Pu"awska et al. / Systematic <strong>and</strong> Applied Microbiology 29 (2006) 470–479providing bacterial strains. J. Pu"awska is deeply indebtedto the Foundation for Polish Science for NationalScholarship for Young Scientist (Edition 2001/2002).A. Willems is grateful to the Fund for ScientificResearch – Fl<strong>and</strong>ers for a postdoctoral research fellowship.This work was supported by Polish Scientific Committee(KBN) Grant 5P06A02119.References[1] J. Brosius, T.J. Dull, D.D. Sleeter, H.F. Noller, Geneorganization <strong>and</strong> primary structure <strong>of</strong> a ribosomal RNAoperon from Escherichia coli, J. Mol. Biol. 148 (1981)107–127.[2] J. Cubero, M.M. Lopez, An efficient microtiter system todetermine <strong>Agrobacterium</strong> biovar, Eur. J. Plant Pathol. 107(2001) 757–760.[3] P. de Lajudie, E. Laurent-Fulele, A. Willems, U. Torck,R. Coopman, M.D. Collins, K. Kersters, B. Dreyfus, M.Gillis, Allorhizobium undicola gen. nov., sp. nov., nitrogen-fixingbacteria that efficiently nodulate Neptunianatans in Senegal, Int. J. Syst. Bacteriol. 48 (1998)1277–1290.[4] P. de Lajudie, A. Willems, B. Pot, D. Dewettinck,G. Maestrojuan, M. Neyra, M.D. Collins, B. Dreyfus,K. Kersters, M. Gillis, Polyphasic taxonomy <strong>of</strong> rhizobia:emendation <strong>of</strong> the genus Sinorhizobium <strong>and</strong> description <strong>of</strong>Sinorhizobium meliloti comb. nov., Sinorhizobium sahelisp. nov., <strong>and</strong> Sinorhizobium teranga sp. nov., Int. J. Syst.Bacteriol. 44 (1994) 715–733.[5] J. De Ley, R. Tijtgat, J. De Smedt, M. Michiels,Thermal stability <strong>of</strong> DNA:DNA hybrids within thegenus <strong>Agrobacterium</strong>, J. Gen. Microbiol. 78 (1973)241–252.[6] X. Dong, G. Cheng, W. Jian, Simultaneous <strong>identification</strong><strong>of</strong> five Bifidobacterium species isolated from human beingsusing multiple PCR primers, Syst. Appl. Microbiol. 23(2000) 386–390.[7] S.K. Farr<strong>and</strong>, P.B. van Berkum, P. Oger, <strong>Agrobacterium</strong>is a definable genus <strong>of</strong> the family Rhizobiaceae, Int.J. Syst. Evol. Microbiol. 53 (2003) 1681–1687.[8] B. Holmes, Taxonomy <strong>of</strong> <strong>Agrobacterium</strong>, Acta Hortic.225 (1988) 47–52.[9] B. Holmes, P. Roberts, The classification, <strong>identification</strong><strong>and</strong> nomenclature <strong>of</strong> agrobacteria, J. Appl. Bacteriol. 50(1981) 443–467.[10] J.G. Holt, N.R. Krieg, P.H.A. Sneath, J.T. Staley, S.T.Williams, Bergey’s Manual <strong>of</strong> Determinative Bacteriology,ninth ed., Williams & Wilkins Comp., Baltimore,MD, USA, 1994.[11] B.D.W. Jarvis, S. Sivakumaran, S.W. Tighe, M. Gillis,Identification <strong>of</strong> <strong>Agrobacterium</strong> <strong>and</strong> Rhizobium speciesbased on cellular fatty acid composition, Plant Soil 184(1996) 143–158.[12] A. Kerr, P. Manigault, J. Tempé, Transfer <strong>of</strong> virulence invivo <strong>and</strong> in vitro in <strong>Agrobacterium</strong>, Nature 265 (1977)560–561.[13] K. Kersters, J. De Ley, P.H.A. Sneath, M. Sackin,Numerical taxonomic analysis <strong>of</strong> <strong>Agrobacterium</strong>, J. Gen.Microbiol. 78 (1973) 227–239.[14] E.O. King, M.K. Ward, D.E. Raney, Two simple mediafor the demonstration <strong>of</strong> pyocyanin <strong>and</strong> fluorescein,J. Lab. Med. 44 (1954) 693–695.[15] C.P. Kolbert, D.H. Persing, Ribosomal DNA sequencingas a tool for <strong>identification</strong> <strong>of</strong> bacterial pathogens, Curr.Opin. Microbiol. 2 (1999) 299–305.[16] U.K. Laemmli, Cleavage <strong>of</strong> structural proteins duringassembly <strong>of</strong> the head <strong>of</strong> bacteriophage T4, Nature 227(1970) 680–685.[17] W. Ludwig, S. Dorn, N. Springer, G. Kirchh<strong>of</strong>, K.H.Schleifer, PCR-based preparation <strong>of</strong> 23S rRNA-targetedgroup-<strong>specific</strong> polynucleotide probes, Appl. Environ.Microbiol. 60 (1994) 3234–3244.[18] L.W. Moore, C.I. Kado, H. Bouzar, II Gram-negativebacteria A, <strong>Agrobacterium</strong>, In: N.W. Schaad (Ed.),Laboratory Guide for Identification <strong>of</strong> Plant PathogenicBacteria, American Phytopathological Society, St. Paul,MN, 1988, pp. 16–36.[19] C. Mougel, B. Cournoyer, X. Nesme, Novel telluriteamendedmedia <strong>and</strong> <strong>specific</strong> chromosomal <strong>and</strong> Ti plasmidprobes for direct analysis <strong>of</strong> soil populations <strong>of</strong> <strong>Agrobacterium</strong>biovars 1 <strong>and</strong> 2, Appl. Environ. Microbiol. 67(2001) 65–74.[20] J. Peplies, F.O. Glo¨ckner, R. Amann, W. Ludwig,Comparative sequence analysis <strong>and</strong> oligonucleotide probedesign based on 23S rRNA genes <strong>of</strong> Alphaproteobacteriafrom North Sea bacterioplankton, Syst. Appl. Microbiol.27 (2004) 573–580.[21] J. Pu"awska, Z. Piotrowska-Seget, Characterization <strong>of</strong><strong>Agrobacterium</strong> isolates from chrysanthemum on the basis<strong>of</strong> biochemical tests, FAME analysis <strong>and</strong> RAPD,Phytopathol. Pol. 30 (2003) 9–17.[22] J. Pu"awska, P. Sobiczewski, Development <strong>of</strong> a seminestedPCR based method for sensitive detection <strong>of</strong>tumorigenic <strong>Agrobacterium</strong> in soil, J. Appl. Microbiol. 98(2005) 710–721.[23] J. Pu"awska, M. Maes, A. Willems, P. Sobiczewski,Phylogenetic analysis <strong>of</strong> 23S rRNA gene sequences <strong>of</strong><strong>Agrobacterium</strong>, Rhizobium <strong>and</strong> Sinorhizobium strains,Syst. Appl. Microbiol. 23 (2000) 238–244.[24] J. Sambrook, E.F. Fritsch, T. Maniatis, MolecularCloning, A Laboratory Manual, second ed., Cold SpringHarbor Laboratory Press, Cold Spring Harbor, NY,1989.[25] H. Sawada, H. Ieki, Phenotypic characteristics <strong>of</strong> thegenus <strong>Agrobacterium</strong>, Ann. Phytopathol. Soc. Jpn. 58(1992) 37–45.[26] H. Sawada, H. Ieki, H. Oyaizu, S. Matsumoto, Proposalfor rejection <strong>of</strong> <strong>Agrobacterium</strong> tumefaciens <strong>and</strong> reviseddescription for the genus <strong>Agrobacterium</strong> <strong>and</strong> for <strong>Agrobacterium</strong>radiobacter <strong>and</strong> <strong>Agrobacterium</strong> rhizogenes, Int.J. Syst. Bacteriol. 43 (1993) 694–702.[27] H. Sawada, Y. Takikawa, H. Ieki, Fatty acid methyl esterpr<strong>of</strong>iles <strong>of</strong> the genus <strong>Agrobacterium</strong>, Ann. Phytopathol.Soc. Jpn. 58 (1992) 46–51.[28] P. Sobiczewski, Etiology <strong>of</strong> crown gall on fruit trees inPol<strong>and</strong>, J. Fruit Ornam. Plant Res. 4 (1996) 147–161.