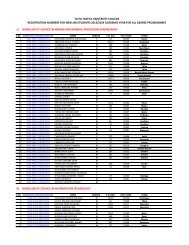
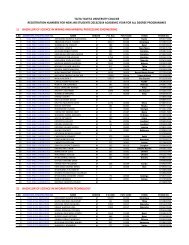
Conference Book of Abstracts - Taita Taveta University College
Conference Book of Abstracts - Taita Taveta University College
Conference Book of Abstracts - Taita Taveta University College
- No tags were found...
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
The Soda Lakes <strong>of</strong> Kenya have attracted much interest from researchers over the years. They areamong the most productive ecosystems in the world with a primary productivity <strong>of</strong> 10g cm -2 day -1 and are very rich in microbial diversity. Due to the highly extreme conditions <strong>of</strong> pH,temperature and salinity, microorganisms from soda lakes have great promise for exploitation inBiotechnology. Using a combination <strong>of</strong> culture dependent and culture independent techniques,the microbial diversity <strong>of</strong> several soda lakes was explored. Clone Library studies <strong>of</strong> LakeElmenteita using bacterial and archeal primers revealed major groups <strong>of</strong> bacteria and archaea. Aclone library <strong>of</strong> bacterial 16S rRNA genes revealed the presence <strong>of</strong> 37 orders in the DomainBacteria. Cyanobacteria were the most abundant clones followed by the phylum Firmicutesgroup. The firmicutes had the most diverse genera represented. All clones affiliated to the classBetaproteobacteria originated from DNA obtained from the water samples. BLAST analysisshowed that 93.1% <strong>of</strong> the sequenced clones had similarity values below 98% to both culturedand as yet uncultured bacteria. Comparative sequence analysis <strong>of</strong> archaeal clones affiliated themto a wide range <strong>of</strong> genera including Natronococcus, Halovivax, Halobiforma, Halorubrum, andHalalkalicoccus. The highest percentage (46%) <strong>of</strong> the clones, however, belonged to unculturedmembers <strong>of</strong> the Domain Archaea in the order Halobacteriales. In a recent study, water, wetsediments, dry sediments, mats and grassland soil were collected at different sampling pointsfrom lakes Bogoria, Magadi, Crater lake Sonachi and Elmenteita in Kenya. The V3 to V5 region<strong>of</strong> the 16S rRNA gene was analyzed using 454 pyrosequencing-based approach to assess thebacterial and archaea diversity in the collected samples. Within the domain Bacteria, a total <strong>of</strong> 47phyla were detected. A total <strong>of</strong> 198 cultured genera were represented in the bacteria dataset. Inthe domain Archaea, the Crenarchaeota and Euryarchaeota were predominant. The PhylumKorarchaeota was detected only in the dry sediments from Lake Bogoria (sample Arc-17B). In13 samples, the Halobacteria were the most abundant while classes Thermoplasmata andMethanomicrobia. A total <strong>of</strong> 30 cultured genera were represented in this dataset. Rarefactionanalysis shows that the water samples and the samples from Lake Magadi were the least diversein terms <strong>of</strong> the number <strong>of</strong> species. The low diversity in the Magadi samples may be attributed tothe high salt and pH which may promote growth <strong>of</strong> specific group <strong>of</strong> organisms that haveadapted to the harsh conditions. Isolation <strong>of</strong> bacteria and fungi from Lake Magadi, Elmenteitaand Nakuru led to the recovery <strong>of</strong> previously isolated as well as yet unrecovered groups. Novelspecies are in the process <strong>of</strong> being described. The results show that the archaeal and Bacterialdiversity in the Soda Lakes could be higher than previously reported.ReferencesMwirichia, R, Cousin, S; Muigai, AW, Boga HI and Stackebrandt, E (2010) Bacterial Diversity in the HaloalkalineLake Elmenteita, Kenya. Curr. Microbiol. 62(1):209-21Mwirichia, R, Muigai, AW, Tindall, B, Boga HI and Stackebrandt, E (2010) Isolation and characterization <strong>of</strong>bacteria from the haloalkaline Lake Elmenteita. Extremophiles 14(4):339-48Mwirichia, R, Cousin, S; Muigai, AW, Boga HI and Stackebrandt, E (2010): Archaeal Diversity in the HaloalkalineLake Elmenteita in Kenya. Curr. Microbiol. 60:47–52Kambura AK (2011) Isolation,characterization and screening <strong>of</strong> bacterial isolates from lake Magadi for Exoenzymesand Antimicrobial Activity. MSc Thesis, Jomo Kenyatta <strong>University</strong> <strong>of</strong> Agriculture and Technology, Kenya