Ca - LISM
Ca - LISM
Ca - LISM
- No tags were found...
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
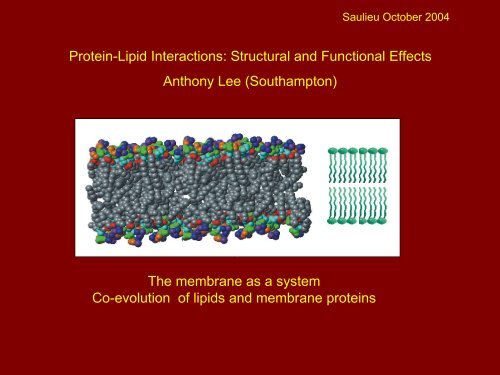
Saulieu October 2004Protein-Lipid Interactions: Structural and Functional EffectsAnthony Lee (Southampton)The membrane as a systemCo-evolution of lipids and membrane proteins
OOOOROPO-OOR+Me 3 N-OO+N H 3HPhosphatidylcholinePhosphatidylserinePhosphatidic acid
Lipid as solventBacteriorhodopsinLipid as cofactorK + channel KcsAhydrophobiccoreAnionic lipidBoundary or Annular Lipid‘Cofactor ‘ or non-annular lipid
Reconstitution of Membrane ProteinsSolubilizationDetergentReconstitutionDetergent RemovalDilutionMembraneFragmentsExcess lipidDialysisBiobeadsSealedvesicles
The importance of Annular LipidThe Lipid Headgroup RegionThe Hydrophobic coreHydrophobic thicknessThe importance of non-annular lipid
<strong>Ca</strong>-ATPase from skeletal muscleSarcoplasmicReticulumNerveDepolarisation Muscle CellSynapsePlasma membrane<strong>Ca</strong> 2+ <strong>Ca</strong> 2+ T system<strong>Ca</strong> 2+2+<strong>Ca</strong> channel<strong>Ca</strong> 2+ <strong>Ca</strong> 2+TnCATP ADP + P iActinMyosin
Effect of Lipid headgroup<strong>Ca</strong>lcium ATPaseSkeletal musclesarcoplasmic reticulumOORP OOOOOATPADP + P i
Relative ATPase Activity10.80.60.40.2<strong>Ca</strong>lcium ATPaseFatty Acyl Chains = Oleoyl (C18:1)SR:70% PC20% PE10% PS/PI0PC PE PSPASR
The importance of Annular LipidThe Lipid Headgroup RegionWhy is activity headgroup dependant?Why does the SR contain anionic lipidsthat support low ATPase activity?The Hydrophobic coreHydrophobic thicknessThe importance of non-annular lipid
<strong>Ca</strong>lcium-ATPaseE2Tg E1<strong>Ca</strong> 2HeadgroupregionR63M1hydrophobiccoreHeadgroupregionR63M1
The importance of Annular LipidThe Lipid Headgroup RegionWhy is activity headgroup dependant?Why does the SR contain anionic lipidsthat support low ATPase activity?The Hydrophobic coreHydrophobic thicknessThe importance of non-annular lipid
<strong>Ca</strong>lcium ATPase activityFractional activity1.00.80.60.40.20.00.0 0.2 0.4 0.6 0.8 1.0Mole Fraction anionic lipiddi(C18:1)PCSR:70% PC20% PE10% PS/PIdi(C18:1)PSdi(C18:1)PA
<strong>Ca</strong>lcium accumulation (nmoles/mg protein)Accumulation of calcium by reconstituted<strong>Ca</strong>-ATPase in sealed vesicles5000400030002000100000 500 1000Time (secs)PC + 10%cardiolipinPC + 10% PAPC<strong>Ca</strong>ATPhydrolysisxleak
<strong>Ca</strong> out<strong>Ca</strong> inE1E2Slippage and Passive Leak<strong>Ca</strong> outE1<strong>Ca</strong> 2E2P iE2PE1<strong>Ca</strong> ATP2<strong>Ca</strong> inE2P<strong>Ca</strong> 2SlippagePassive leaktransportmM <strong>Ca</strong>ATPhydrolysispassiveleakslippage<strong>Ca</strong> outPassive LeakSlippage6000<strong>Ca</strong> accumulated(nmoles/mg)5000400030002000100000 200 400 600 800 1000Time (sec)0 200 400 600 800 1000Time (sec)
<strong>Ca</strong> outSlippage and LeakSlippageATPhydrolysis<strong>Ca</strong> out<strong>Ca</strong> inE1E2E1<strong>Ca</strong> 2E2P iE2PE1<strong>Ca</strong> ATP2<strong>Ca</strong> inE2P<strong>Ca</strong> 2transportmM <strong>Ca</strong>passiveleakslippage<strong>Ca</strong> outSlippage<strong>Ca</strong> accumulated(nmoles/mg)50004000300020001000PC + 10%cardiolipinIncreasingslippagePC+10%PAPC00 500 1000Time (secs)
inslippagecytoplasmsurfacetransport
SarcoplasmicReticulumNerveDepolarisation Muscle CellSynapsePlasma membrane<strong>Ca</strong> 2+ <strong>Ca</strong> 2+ T system<strong>Ca</strong> 2+2+<strong>Ca</strong> channel<strong>Ca</strong> 2+ <strong>Ca</strong> 2+TnCATP ADP + P ActiniMyosinRole of SR: accumulate <strong>Ca</strong>, leading to muscle relaxationthermogenesis
<strong>Ca</strong> outHeat generationE1E2E1<strong>Ca</strong> 2E2P iE2PE1<strong>Ca</strong> ATP2<strong>Ca</strong> inE2P<strong>Ca</strong> 2transportSlippage = heat generation<strong>Ca</strong> out12001000DOPCFast slippageHeat (mcal/mg)800600400200DOPC +20% DOPASlow slippage00 5 10 15 20 25 30 35ATPTime (min)
Sarcoplasmic reticulum contains sarcolipinMERSTRELCLNFTVVLITVILIWLLVRSYQY<strong>Ca</strong> accumulation (nmoles/mg protein)20001500100050010:120:100 200 400 600 800 1000 1200Time (secs)SLN:ATPase02:15:1ATP hydrolysisslippagemM <strong>Ca</strong>Sarcolipin inceases slippage
SarcoplasmicReticulumNerveDepolarisation Muscle CellSynapsePlasma membrane<strong>Ca</strong> 2+ <strong>Ca</strong> 2+ T system<strong>Ca</strong> 2+2+<strong>Ca</strong> channel<strong>Ca</strong> 2+ <strong>Ca</strong> 2+TnCATP ADP + P iActinMyosinRoles of SR: accumulate <strong>Ca</strong>ThermogenesisSarcolipinAnionic Lipid
The importance of Annular LipidThe Lipid Headgroup RegionImportant for functionNature of binding siteon proteinThe Hydrophobic coreHydrophobic thicknessThe importance of non-annular lipid
Lipid BindingSpecificNon-specificY39LysG256K35PCTrpPCR31PEPhotosynthetic reactioncentreMD simulation ofhelix in PC bilayer(M. Sansom)
The importance of Annular LipidThe Lipid Headgroup RegionImportant for functionThe Hydrophobic coreHydrophobic thicknessThe importance of non-annular lipid
How efficient is hydrophobic matching between the bilayer anda membrane protein ?The use of Trp asa reporter grouppolarbacteriorhodopsinhydrophobic
How efficient is hydrophobic matching?Trp fluorescence spectraW1.5Intensity1.00.5FreeTrp0.0300 320 340 360 380 400Wavelength (nm)
K + channel KcsA – How efficient is hydrophobic matching27 ÅC1816 ÅC1038 ÅC24
Reconstituted KcsAPhosphatidylcholineChain Length1.4 C101.21.00.80.60.40.20.0FreeTrpC18C24-0.2300 320 340 360 380 400Wavelength (nm)
Tryptophan residues do not change their environmentwith changing bilayer thicknessC18C10C24Hydrophobic matching is highly efficientEither lipid distorts to match proteinor protein distorts to match lipidor both
Hydrophobic MismatchBilayer Distortion to Match ProteinBilayer too thickCompressHydrophobicthicknessWorkOptimalmatchingStretchBilayer too thin
Fluorescence QuenchingShort range – requires contact between fluorophore and quencherBrOORP OOOBrBrOOBrexcitinglightexcitinglight*excitedstateTrpheat100 %*heatemission offluorescence lightQuencher (Br)
Fluorescence quenchingin mixtures of brominated and non-brominated lipidOORP OOOOORO P OOOOOOBr-Lipid matchesHigh quenchingWWBrBrW W WBrBrHydrophobicmatchingNon-Br-LipidmatchesLow quenchingWWW WW
Binding of lipid to a membrane proteinExchange at a set of annular sitesBA1.00.80.6Prot.A + BKProt.B + AF/F o0.40.20.00.0 0.2 0.4 0.6 0.8 1.0Mole Fraction Brominated PhospholipidK is binding constant for B relative to A[A is di(Br 2C18:0)PC]
Relative binding constant2.01.51.00.5Relative Lipid Binding Constants[relative to di(Br 2C18:0)PC]0.08 10 12 14 16 18 20 22 24 26Fatty Acyl Chain LengthOmpFKcsA34 Å
Hydrophobic mismatch – protein distortionChange in helix tilt: changes at the ends of the helicesThick bilayerThin bilayerChanges in heliceschange in protein activity
R<strong>Ca</strong>lcium ATPaseChangingBilayer thicknessOP OOOOOODistortion ofproteinChange inactivityATPADP + P i
42+<strong>Ca</strong>- ATPasePCATPase activity (IU/mg)321Me 3 N +OOPO-OOOOO012 14 16 18 20 22 24Fatty acyl chain
Diacylglycerol kinase of E. coliPeriplasmCytoplasmDiacylglycerol + ATPPhosphatidic acid + ADP
Diacylglycerol kinaseDihexanoylglycerol + ATPPhosphatidic acid + ADPActivity (IU/mg)604020PC014 16 18 20 22 24Chain length
Hydrophobic mismatch<strong>Ca</strong>lcium ATPaseChange in helix tiltChanges at ends ofhelicesChange in helix packing
Helix-Helix InteractionsKcsA channelL105V106A29Knobs-into-hole packing
Fluorescence quenchingTrp containing peptideDibromo-tyrosine containing peptideW Q Dibromo-Tyr1.00.80.6PCC14Dimer formationFlu. quenchFluorescence0.40.2C18C24W Q0.00 0.02 0.04Mole fraction brominated peptide
Unitary free energies of helix-helix interactionG o = -RT ln KG o (kJ mol -1 )1210864200.5 kJ mol -1 per C atomL 22-Q 220 4 8 12 16 20 24Fatty acyl chain lengthW Q Dibromo-TyrDimer formationFlu. quenchW Q
Hydrophobic matching between the bilayer anda membrane protein is very efficientHow can we measure the hydrophobic thickness ofa membrane protein?<strong>Ca</strong>n we use Trpmutagenesis +Fluorescence?polarhydrophobicbacteriorhodopsin
Do Trp residues anchor transmembrane α-helicesinto the membrane?NTransmembrane domainTrp Ile ValLeuTyrTrpPheCKcsA
Trp at ends of helices are not conservedKcsAPore of Kv
Effects of Trp mutagenesis on activityDiacylglycerol kinase of E. coliPeriplasm*48 52W47*W117W112CytoplasmW18 W25*29Diacylglycerol + ATP68 96* **charged residuesPhosphatidic acid + ADP
Use of Trp fluorescence to define hydrophobic coreof the mechanosensitive channel of large conductance MscLD68D36periplasmL69M2F80D16L92cytoplasm
Use of Trp fluorescence to define hydrophobic coreof MscLEmission Max344340336332328324external residuesDOPC69 92interface32051 66 69 72 75 78 81 84 87 90 93 96 99 102105Residue Number
D6825 A O L69V91L92R11Y94D16
The importance of Annular LipidThe Lipid Headgroup RegionImportant for functionThe Hydrophobic coreHydrophobic thicknessThe importance of non-annular lipid
KcsAnon-annularlipidanionicannularlipidAnionic lipid essential for activityIn the absence of anionic lipid is the non-annular site empty?
AnioniclipidKcsAR o= 8Å3 Trpnot quenchedW682 TrpK +W67non-annularquenchedW87annularquenchednonannularlipid
KcsA Trp quenchingnon-annularoF/F1.00.90.80.70.6DOPC/BrPCDOPC/BrPA0.50.0 0.2 0.4 0.6 0.8 1.0Brominated PhospholipidannularW87(annular)W87+W67(annular + non-annular)The non-annular site can only be occupied by anionic lipid
KcsAR64R89Anioniclipid
Lipid-Protein InteractionsAnnular lipidThickness:Lipid headgroup:tilt of helices/helix endspacking of helicesmolecular interactionsNonannular lipidHigh specificityLipid ‘co-factor’
Lipid-Protein InteractionsMalcolm EastKcsA:Ian WilliamsonSimon AlvisMscL:Andy PowlDiacylglycerolkinaseElizabeth Clark<strong>Ca</strong> 2+ -ATPaseAnthony StarlingSanja MallOmpF:Aisling O’KeeffeModel HelicesSanja Mall