here - Biomedical Computation Review
here - Biomedical Computation Review
here - Biomedical Computation Review
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
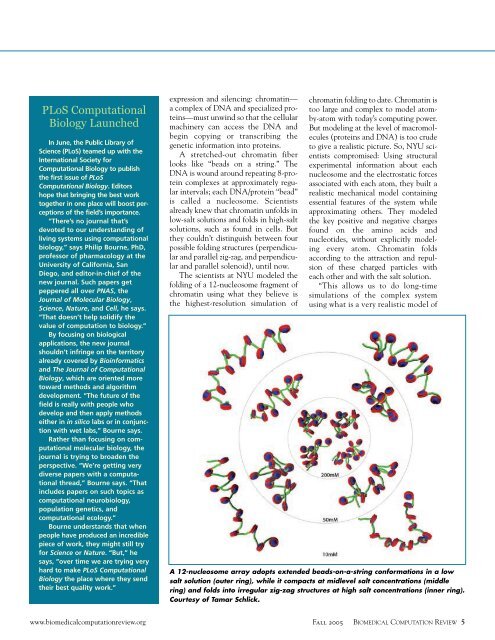
PLoS <strong>Computation</strong>alBiology LaunchedIn June, the Public Library ofScience (PLoS) teamed up with theInternational Society for<strong>Computation</strong>al Biology to publishthe first issue of PLoS<strong>Computation</strong>al Biology. Editorshope that bringing the best worktogether in one place will boost perceptionsof the field’s importance.“T<strong>here</strong>’s no journal that’sdevoted to our understanding ofliving systems using computationalbiology,” says Philip Bourne, PhD,professor of pharmacology at theUniversity of California, SanDiego, and editor-in-chief of thenew journal. Such papers getpeppered all over PNAS, theJournal of Molecular Biology,Science, Nature, and Cell, he says.“That doesn’t help solidify thevalue of computation to biology.”By focusing on biologicalapplications, the new journalshouldn’t infringe on the territoryalready covered by Bioinformaticsand The Journal of <strong>Computation</strong>alBiology, which are oriented moretoward methods and algorithmdevelopment. “The future of thefield is really with people whodevelop and then apply methodseither in in silico labs or in conjunctionwith wet labs,” Bourne says.Rather than focusing on computationalmolecular biology, thejournal is trying to broaden theperspective. “We’re getting verydiverse papers with a computationalthread,” Bourne says. “Thatincludes papers on such topics ascomputational neurobiology,population genetics, andcomputational ecology.”Bourne understands that whenpeople have produced an incrediblepiece of work, they might still tryfor Science or Nature. “But,” hesays, “over time we are trying veryhard to make PLoS <strong>Computation</strong>alBiology the place w<strong>here</strong> they sendtheir best quality work.”expression and silencing: chromatin—a complex of DNA and specialized proteins—mustunwind so that the cellularmachinery can access the DNA andbegin copying or transcribing thegenetic information into proteins.A stretched-out chromatin fiberlooks like “beads on a string.” TheDNA is wound around repeating 8-proteincomplexes at approximately regularintervals; each DNA/protein “bead”is called a nucleosome. Scientistsalready knew that chromatin unfolds inlow-salt solutions and folds in high-saltsolutions, such as found in cells. Butthey couldn’t distinguish between fourpossible folding structures (perpendicularand parallel zig-zag, and perpendicularand parallel solenoid), until now.The scientists at NYU modeled thefolding of a 12-nucleosome fragment ofchromatin using what they believe isthe highest-resolution simulation ofchromatin folding to date. Chromatin istoo large and complex to model atomby-atomwith today’s computing power.But modeling at the level of macromolecules(proteins and DNA) is too crudeto give a realistic picture. So, NYU scientistscompromised: Using structuralexperimental information about eachnucleosome and the electrostatic forcesassociated with each atom, they built arealistic mechanical model containingessential features of the system whileapproximating others. They modeledthe key positive and negative chargesfound on the amino acids andnucleotides, without explicitly modelingevery atom. Chromatin foldsaccording to the attraction and repulsionof these charged particles witheach other and with the salt solution.“This allows us to do long-timesimulations of the complex systemusing what is a very realistic model ofA 12-nucleosome array adopts extended beads-on-a-string conformations in a lowsalt solution (outer ring), while it compacts at midlevel salt concentrations (middlering) and folds into irregular zig-zag structures at high salt concentrations (inner ring).Courtesy of Tamar Schlick.www.biomedicalcomputationreview.orgFall 2005 BIOMEDICAL COMPUTATION REVIEW 5