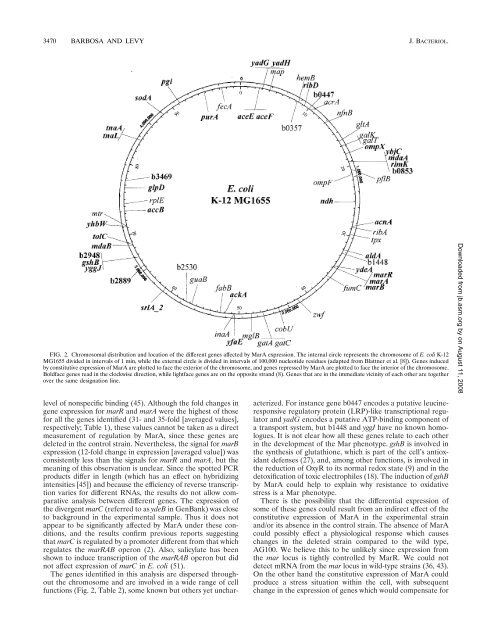
3470 BARBOSA AND LEVY J. BACTERIOL.FIG. 2. <strong>Chromosomal</strong> distribution and location <strong>of</strong> the different genes affected by MarA expression. The <strong>in</strong>ternal circle represents the chromosome <strong>of</strong> E. coli K-12MG1655 divided <strong>in</strong> <strong>in</strong>tervals <strong>of</strong> 1 m<strong>in</strong>, while the external circle is divided <strong>in</strong> <strong>in</strong>tervals <strong>of</strong> 100,000 nucleotide residues (adapted from Blattner et al. [8]). <strong>Genes</strong> <strong>in</strong>ducedby constitutive expression <strong>of</strong> MarA are plotted to face the exterior <strong>of</strong> the chromosome, and genes repressed by MarA are plotted to face the <strong>in</strong>terior <strong>of</strong> the chromosome.Boldface genes read <strong>in</strong> the clockwise direction, while lightface genes are on the opposite strand (8). <strong>Genes</strong> that are <strong>in</strong> the immediate vic<strong>in</strong>ity <strong>of</strong> each other are together<strong>over</strong> the same designation l<strong>in</strong>e.level <strong>of</strong> nonspecific b<strong>in</strong>d<strong>in</strong>g (45). Although the fold changes <strong>in</strong>gene expression for marR and marA were the highest <strong>of</strong> thosefor all the genes identified (31- and 35-fold [averaged values],respectively; Table 1), these values cannot be taken as a directmeasurement <strong>of</strong> regulation by MarA, s<strong>in</strong>ce these genes aredeleted <strong>in</strong> the control stra<strong>in</strong>. Nevertheless, the signal for marBexpression (12-fold change <strong>in</strong> expression [averaged value]) wasconsistently less than the signals for marR and marA, but themean<strong>in</strong>g <strong>of</strong> this observation is unclear. S<strong>in</strong>ce the spotted PCRproducts differ <strong>in</strong> length (which has an effect on hybridiz<strong>in</strong>g<strong>in</strong>tensities [45]) and because the efficiency <strong>of</strong> reverse transcriptionvaries for different RNAs, the results do not allow comparativeanalysis between different genes. The expression <strong>of</strong>the divergent marC (referred to as ydeB <strong>in</strong> GenBank) was closeto background <strong>in</strong> the experimental sample. Thus it does notappear to be significantly affected by MarA under these conditions,and the results confirm previous reports suggest<strong>in</strong>gthat marC is regulated by a promoter different from that whichregulates the marRAB operon (2). Also, salicylate has beenshown to <strong>in</strong>duce transcription <strong>of</strong> the marRAB operon but didnot affect expression <strong>of</strong> marC <strong>in</strong> E. coli (51).The genes identified <strong>in</strong> this analysis are dispersed throughoutthe chromosome and are <strong>in</strong>volved <strong>in</strong> a wide range <strong>of</strong> cellfunctions (Fig. 2, Table 2), some known but others yet uncharacterized.For <strong>in</strong>stance gene b0447 encodes a putative leuc<strong>in</strong>eresponsiveregulatory prote<strong>in</strong> (LRP)-like transcriptional regulatorand yadG encodes a putative ATP-b<strong>in</strong>d<strong>in</strong>g component <strong>of</strong>a transport system, but b1448 and yggJ have no known homologues.It is not clear how all these genes relate to each other<strong>in</strong> the development <strong>of</strong> the Mar phenotype. gshB is <strong>in</strong>volved <strong>in</strong>the synthesis <strong>of</strong> glutathione, which is part <strong>of</strong> the cell’s antioxidantdefenses (27), and, among other functions, is <strong>in</strong>volved <strong>in</strong>the reduction <strong>of</strong> OxyR to its normal redox state (9) and <strong>in</strong> thedetoxification <strong>of</strong> toxic electrophiles (18). The <strong>in</strong>duction <strong>of</strong> gshBby MarA could help to expla<strong>in</strong> why resistance to oxidativestress is a Mar phenotype.There is the possibility that the differential expression <strong>of</strong>some <strong>of</strong> these genes could result from an <strong>in</strong>direct effect <strong>of</strong> theconstitutive expression <strong>of</strong> MarA <strong>in</strong> the experimental stra<strong>in</strong>and/or its absence <strong>in</strong> the control stra<strong>in</strong>. The absence <strong>of</strong> MarAcould possibly effect a physiological response which causeschanges <strong>in</strong> the deleted stra<strong>in</strong> compared to the wild type,AG100. We believe this to be unlikely s<strong>in</strong>ce expression fromthe mar locus is tightly controlled by MarR. We could notdetect mRNA from the mar locus <strong>in</strong> wild-type stra<strong>in</strong>s (36, 43).On the other hand the constitutive expression <strong>of</strong> MarA couldproduce a stress situation with<strong>in</strong> the cell, with subsequentchange <strong>in</strong> the expression <strong>of</strong> genes which would compensate forDownloaded from jb.asm.org by on August 11, 2008
VOL. 182, 2000 EXPRESSION PROFILING OF THE E. COLI mar REGULON 3471TABLE 2. Functional classification <strong>of</strong> genes affected byMarA expressionPhysiological function a<strong>Genes</strong>Energy metabolism, carbon ................aceE, aceF, ackA, acnA, aldA, fumC,glpD, gltA, mdaA, ndh, pflB, pgi,zwfBiosynthesis <strong>of</strong> c<strong>of</strong>actors, carriers......accB, cobU, hemB, gshB, ribA, ribDCarbon compound catabolism............galK, galTAm<strong>in</strong>o acid biosynthesis andmetabolism........................................tnaA, tnaLFatty acid biosynthesis.........................fabBNucleotide biosynthesis .......................guaB, purAAdaptation ............................................<strong>in</strong>aA bTransport/b<strong>in</strong>d<strong>in</strong>g prote<strong>in</strong>s..................gatA, gatC, fecA, mglB, mtr, srlA2,tolC c , yadG, yadH, ydeA, b3469Protection responses............................acrA, marA, marB, marR, nfnB, sodA,tpxCell envelope ........................................ompF, ompXRibosome constituents.........................rimK, rplEMacromolecule synthesis,modification ......................................mapNot classified.........................................b0357, b0447, b0853, mdaB, yhbWEncod<strong>in</strong>g unknown prote<strong>in</strong>s ...............b1448, b2530, b2889, b2948, ybjC,yfaE, yggJa From GenProtEC E. coli genome and proteome database (http://genprotec.mbl.edu/start).b Based on presumptive evidence.c Also <strong>in</strong>volved <strong>in</strong> cell division and protection responses.the possible adverse effects. Nevertheless, naturally occurr<strong>in</strong>gmar mutants among cl<strong>in</strong>ical isolates <strong>of</strong> E. coli (36, 43) which,like our experimental stra<strong>in</strong>, constitutively express MarA havebeen reported. Still, differences <strong>in</strong> the quantity <strong>of</strong> MarA, i.e.,produced with low-copy-number vector pMAK705 versus s<strong>in</strong>gle-copymarA on the chromosome, may <strong>in</strong>fluence the results.Although no <strong>in</strong>-depth comparative physiological studieswere carried out, no difference <strong>in</strong> growth rate between the wildtype (AG100) and the mar-deleted stra<strong>in</strong> (AG100Kan) wasfound. Control stra<strong>in</strong> AG100Kan carry<strong>in</strong>g pMAK705 had agrowth rate 6% slower than that <strong>of</strong> AG100 or AG100Kan, andthe experimental stra<strong>in</strong>, AG100Kan[pAS10], had a growth ratewhich was 15% slower than that <strong>of</strong> the control stra<strong>in</strong>. Whilethis growth difference could possibly affect the expression <strong>of</strong>some <strong>of</strong> the reported genes, an effect on the bacterial growthwould not be unexpected as an <strong>in</strong>tegral part <strong>of</strong> a stress responsesystem such as mar.Despite the fact that AG100Kan[pAS10] constitutively expressesboth MarA and MarB prote<strong>in</strong>s, we believe the differentialregulation <strong>of</strong> the multiple genes here reported to be associatedwith MarA, as MarB shows no characteristics <strong>of</strong> atranscriptional activator. Additionally marB does not appear tobe necessary either for basal or <strong>in</strong>ducible expression <strong>of</strong> the marregulon or for the selection <strong>of</strong> mar mutants (39). Neverthelesswe cannot rule out the possibility that MarB may <strong>in</strong>directlyaffect the expression <strong>of</strong> some <strong>of</strong> these genes, e.g., by trigger<strong>in</strong>ga non-mar-regulated response <strong>in</strong> the cell.Confirmation <strong>of</strong> previously identified MarA-regulated genes.The differential expression <strong>of</strong> most <strong>of</strong> the genes previouslyidentified as part <strong>of</strong> the mar regulon, e.g., <strong>in</strong>aA, sodA, ompF,zwf, and fumC (4, 25, 30, 46) was confirmed (Table 1). A majorrole <strong>in</strong> the Mar phenotype is played by the efflux system acrAB,which acts by pump<strong>in</strong>g toxic compounds out <strong>of</strong> the cell (42, 44,54). An <strong>in</strong>crease <strong>in</strong> the expression <strong>of</strong> the acrA gene <strong>of</strong> theacrAB operon was also observed (Table 1); however, the expressionvalues for acrB were not above background. This k<strong>in</strong>d<strong>of</strong> f<strong>in</strong>d<strong>in</strong>g is not fully understood but could arise from differentialprocess<strong>in</strong>g <strong>of</strong> the polycistronic transcript and/or by differences<strong>in</strong> transcript stability.Previous studies suggest coord<strong>in</strong>ate activation <strong>of</strong> TolC andthe AcrAB efflux pump <strong>in</strong> the development <strong>of</strong> the Mar phenotype(3, 19). Changes <strong>in</strong> the expression <strong>of</strong> outer membraneprote<strong>in</strong>s (e.g., <strong>in</strong>creased OmpX expression and decreasedOmpF and LamB expression) <strong>in</strong> E. coli marR mutants andwild-type stra<strong>in</strong>s <strong>over</strong>express<strong>in</strong>g MarA have also been reported(3, 13). Down-regulation <strong>of</strong> ompF translation is controlled bymicF, a regulatory antisense RNA known to be activated byMarA, which b<strong>in</strong>ds to the 5 untranslated region <strong>of</strong> the formergene mRNA, block<strong>in</strong>g translation (16). We confirm some <strong>of</strong>these reports and show for the first time that MarA expression<strong>in</strong>creases the transcription <strong>of</strong> both tolC and ompX (Table 1).Although we observed a decrease <strong>in</strong> the levels <strong>of</strong> ompF, wefound no evidence for a similar decrease <strong>in</strong> lamB expression,suggest<strong>in</strong>g that LamB may not be the underproduced prote<strong>in</strong>identified <strong>in</strong> the earlier study (3) or that regulation may beposttranscriptional. The micF gene is not spotted on the arrays(which conta<strong>in</strong> only genes cod<strong>in</strong>g for ORFs), and therefore wewere unable to confirm activation <strong>of</strong> this gene by MarA. Nevertheless,under this assumption and given the observed downregulation<strong>of</strong> ompF, the results <strong>in</strong>dicate that micF is also <strong>in</strong>volved<strong>in</strong> the destabilization <strong>of</strong> the ompF mRNA as suggestedby others (13, 16).Transcription <strong>of</strong> the previously identified mlr1 (b1451) andmlr2 (b0<strong>60</strong>3) genes (48) was <strong>in</strong>creased <strong>in</strong> the mar-express<strong>in</strong>gstra<strong>in</strong> <strong>in</strong> two experiments but appeared to be unaffected <strong>in</strong> athird experiment, so these genes were not <strong>in</strong>cluded <strong>in</strong> Table 1.<strong>Expression</strong> <strong>of</strong> the slp gene, previously described as repressedby MarA (48), was so low that any mar-mediated change wouldhave been difficult to detect. This observation may reflect thefact that our experiments were performed on cells <strong>in</strong> mid-logarithmicphase while slp is a stationary-phase-<strong>in</strong>ducible gene (48).Relationship between soxRS and mar regulons. SoxS is theactivator <strong>of</strong> the soxRS regulon (17), which mediates a cellularresponse to oxidative stress and, like MarA, is a member <strong>of</strong> theXylS/AraC family <strong>of</strong> transcriptional activators (20). Many oxidativestress genes, which are known to respond to SoxS, arealso responsive to MarA (30, 41). Conversely, SoxS is able toconfer a Mar phenotype via activation <strong>of</strong> genes that are underthe control <strong>of</strong> MarA (4, 25). <strong>Genes</strong> known to be regulateddirectly or <strong>in</strong>directly by both the MarA and SoxS regulators<strong>in</strong>clude zwf, fpr, fumC, micF, nfo, <strong>in</strong>aA, sodA, and acrAB (4, 25,30, 46, 54). We confirmed the positive regulation <strong>of</strong> zwf, fumC,acrA, <strong>in</strong>aA, and sodA by MarA and also the down-regulation <strong>of</strong>ompF. However, although b<strong>in</strong>d<strong>in</strong>g <strong>of</strong> MarA to nfo and fpr wasshown <strong>in</strong> cell-free studies (30), no significant change <strong>in</strong> expression<strong>of</strong> these two genes was detected us<strong>in</strong>g the experimentalconditions employed here.Our f<strong>in</strong>d<strong>in</strong>gs revealed further <strong>over</strong>lap between the mar andsoxRS regulons. The levels <strong>of</strong> aconitase (acnA) and GTP cyclohydrolaseII (ribA) genes and that <strong>of</strong> the major oxygen<strong>in</strong>sensitivenitroreductase gene (nfsA/mdaA), previously knownto be under the control <strong>of</strong> soxRS (15, 31, 33), were <strong>in</strong>creased <strong>in</strong>mar-express<strong>in</strong>g stra<strong>in</strong>s (Table 1). While NfsA was shown to bethe major isoenzyme affected by paraquat (33), the oxygensensitiveNADPH nitroreductase B gene, nfnB (also designatednfsB), was shown to be slightly <strong>in</strong>duced. We found thatnfnB, like nfsA, is under the positive control <strong>of</strong> MarA (Table1).nfsA was <strong>in</strong>itially designated mdaA (modulator <strong>of</strong> drug activity),as one <strong>of</strong> two genes associated with bacterial resistanceto tumoricidal compounds (10). The other gene, designatedmdaB, was also found to be affected by MarA (Table 1). Informationabout mdaB is very limited, and its function rema<strong>in</strong>sunknown. Our f<strong>in</strong>d<strong>in</strong>gs provide suggestive evidence for a pu-Downloaded from jb.asm.org by on August 11, 2008