PREP Quickstart, Multivariable Calculus Sage Quickstart for ...
PREP Quickstart, Multivariable Calculus Sage Quickstart for ...
PREP Quickstart, Multivariable Calculus Sage Quickstart for ...
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
25, )1254(6,2 t, )34 t2 (2,2/20/13 <strong>PREP</strong> <strong>Quickstart</strong>, <strong>Multivariable</strong> <strong>Calculus</strong> -- <strong>Sage</strong>Toggle Advanced ControlsHelp <strong>for</strong> Jmol 3-D viewerVector-Valued Functionsvar('t')r=vector((2*t-4, t^2, (1/4)*t^3))rr(t=5)(2 t − 4, , ) t 2 1 4 t 3The following makes the derivative also a vector-valued callable expression.velocity = r.diff(t) # velocity(t) = list(r.diff(t)) also wouldworkvelocityvelocity(t=1)test.sagenb.org/home/Carlos_Rodriguez/20/print 2/9(2, 2, )
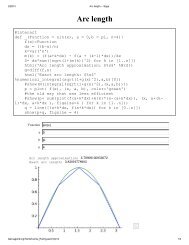
(2, 2, )3 42/20/13 <strong>PREP</strong> <strong>Quickstart</strong>, <strong>Multivariable</strong> <strong>Calculus</strong> -- <strong>Sage</strong>If we know a little vector calculus, we can also do line integrals. Here we compute the arclength betweenand t = π.T=velocity/velocity.norm()t = 0T(t=1).n()(0.683486126173409, 0.683486126173409, 0.256307297315028)We can even then get the arc length by integrating the normalized derivative. As pointed out in the calculustutorial, the syntax <strong>for</strong> numerical_integral is slightly nonstandard - we just put in the endpoints, not the variable.arc_length = numerical_integral(rprime.norm(), 0,1)arc_lengthTraceback (click to the left of this block <strong>for</strong> traceback)...NameError: name 'rprime' is not definedIf we know a little vector calculus, we can also do line integrals. Here we compute a number of quantities.var('x,y,t')density(x,y)=x^2+ytstart=0tend=pit_range=(t,tstart,tend)r_prime = diff(r,t)ds=diff(r,t).norm()arclength=integral(ds,t, tstart, tend)To do line integrals, let's make a function that will calculatevarious differentials to to easily specify the dtintegrands.. We'll use our <strong>for</strong>mulas relating# Let's make a function right off to evaluate an integral overthis curve.# this function uses our values of r, tstart, and tenddef line_integral(integrand):return RR(numerical_integral((integrand).subs(x=r[0],y=r[1],z=r[2]), tstart, tend)[0])var('x,y,z,t')r_prime = diff(r,t)∫ (integrand)dttest.sagenb.org/home/Carlos_Rodriguez/20/print 3/9
2/20/13 <strong>PREP</strong> <strong>Quickstart</strong>, <strong>Multivariable</strong> <strong>Calculus</strong> -- <strong>Sage</strong>ds=diff(r,t).norm()s=line_integral(ds)centroid_x=line_integral(x*ds)/scentroid_y=line_integral(y*ds)/scentroid_z=line_integral(z*ds)/sdm=density(x,y,z)*dsm=line_integral(dm)avg_density = m/smoment_about_yz_plane=line_integral(x*dm)moment_about_xz_plane=line_integral(y*dm)moment_about_xy_plane=line_integral(z*dm)center_mass_x = moment_about_yz_plane/mcenter_mass_y = moment_about_xz_plane/mcenter_mass_z = moment_about_xy_plane/mIx=line_integral((y^2+z^2)*dm)Iy=line_integral((x^2+z^2)*dm)Iz=line_integral((x^2+y^2)*dm)Rx = sqrt(Ix/m)Ry = sqrt(Iy/m)Rz = sqrt(Iz/m)Let's display everything in a nice table.html.table([[r"Density $\delta(x,y)$", density],[r"Curve $\vec r(t)$",r],[r"$t$ range", t_range],[r"$\vec r'(t)$", r_prime],[r"$ds$, a little bit of arclength", ds],[r"$s$ - arclength", s],[r"Centroid (constant density) $\left(\frac{1}{m}\intx\,ds,\frac{1}{m}\int y\,ds, \frac{1}{m}\int z\,ds\right)$",(centroid_x,centroid_y,centroid_z)],[r"$dm=\delta ds$ - a little bit of mass", dm],[r"$m=\int \delta ds$ - mass", m],[r"average density $\frac{1}{m}\int ds$" , avg_density.n()],[r"$M_{yz}=\int x dm$ - moment about $yz$ plane",moment_about_yz_plane],[r"$M_{xz}=\int y dm$ - moment about $xz$ plane",moment_about_xz_plane],[r"$M_{xy}=\int z dm$ - moment about $xy$ plane",moment_about_xy_plane],[r"Center of mass $\left(\frac1m \int xdm, \frac1m \int ydm,\frac1m \int z dm\right)$", (center_mass_x, center_mass_y,test.sagenb.org/home/Carlos_Rodriguez/20/print 4/9
2/20/13 <strong>PREP</strong> <strong>Quickstart</strong>, <strong>Multivariable</strong> <strong>Calculus</strong> -- <strong>Sage</strong>center_mass_z)],[r"$I_x = \int (y^2+z^2) dm$", Ix],[r"$I_y=\int (x^2+z^2) dm$",Iy],[mp(r"$I_z=\int (x^2+y^2)dm$"), Iz],[r"$R_x=\sqrt{I_x/m}$", Rx],[mp(r"$R_y=\sqrt{I_y/m}"), Ry],[mp(r"$R_z=\sqrt{I_z/m}"),Rz]])Traceback (click to the left of this block <strong>for</strong> traceback)...NameError: name 'mp' is not definedWe can also plot vector fields, even in three dimensions.x,y,z=var('x y z')plot_vector_field3d((x*cos(z),-y*cos(z),sin(z)), (x,0,pi),(y,0,pi), (z,0,pi),colors=['red','green','blue'])Toggle Advanced ControlsHelp <strong>for</strong> Jmol 3-D viewerFunctions of Several VariablesThis connects directly to other issues of multivariable functions.How to view these was mostly addressed in the various plotting tutorials. Here is a reminder of what can betest.sagenb.org/home/Carlos_Rodriguez/20/print 5/9
2/20/13 <strong>PREP</strong> <strong>Quickstart</strong>, <strong>Multivariable</strong> <strong>Calculus</strong> -- <strong>Sage</strong>done.# import matplotlib.cm; matplotlib.cm.datad.keys()# 'Spectral', 'summer', 'blues'g(x,y)=e^-x*sin(y)contour_plot(g, (x, -2, 2), (y, -4*pi, 4*pi), cmap = 'Blues',contours=10, colorbar=True)Partial DifferentiationThe following exercise is from Hass, Weir, and Thomas, University <strong>Calculus</strong>, Exercise 12.7.35. It has a localminimum at .(4,−2)f(x, y) = x^2 + x*y + y^2 - 6*x + 2test.sagenb.org/home/Carlos_Rodriguez/20/print 6/9
↦ 2 x + y − 6(x,y)↦ x + 2 y(x,y)f xxD = 3f xxf xx2/20/13 <strong>PREP</strong> <strong>Quickstart</strong>, <strong>Multivariable</strong> <strong>Calculus</strong> -- <strong>Sage</strong>Quiz: Why did we not need to declare the variables in this case?fx(x,y)= f.diff(x)fy(x,y) = f.diff(y)fx; fyf.gradient()solve([fx==0, fy==0], (x, y))(x,y) ↦ (2 x + y − 6, x + 2 y)H = f.hessian()H(x,y)[[x = 4,y = (−2)]]2112And of course if the Hessian has positive determinant andis positive, we have a local minimum.( )Notice how we were able to use many things we've done up to now to solve this.MatricesSymbolic functionsSolvingDifferential calculusSpecial <strong>for</strong>matting commandsAnd, below, plotting!html("$f_{xx}=%s$"%H(4,-2)[0,0])html("$D=%s$"%H(4,-2).det())= 2And of course if the Hessian is positive definite andis positive, we have a local minimum.Notice how we were able to use many things we've done up to now to solve this.test.sagenb.org/home/Carlos_Rodriguez/20/print 7/9
2/20/13 <strong>PREP</strong> <strong>Quickstart</strong>, <strong>Multivariable</strong> <strong>Calculus</strong> -- <strong>Sage</strong>MatricesSymbolic functionsSolvingDifferential calculusSpecial <strong>for</strong>matting commandsAnd, below, plotting!plot3d(f,(x,-5,5),(y,-5,5))+point((4,-2,f(4,-2)),color='red',size=20)Toggle Advanced ControlsHelp <strong>for</strong> Jmol 3-D viewerf(x,y)=x^2*yQuiz: why did we need to declare variables this time?# integrate in the order dy dxf(x,y).integrate(y,0,4*x).integrate(x,0,3)19445# another way to integrate, and in the opposite order toointegrate( integrate(f(x,y), (x, y/4, 3)), (y, 0, 12) )19445var('u v')surface = plot3d(f(x,y), (x, 0, 3.2), (y, 0, 12.3), color =test.sagenb.org/home/Carlos_Rodriguez/20/print 8/9
2/20/13 <strong>PREP</strong> <strong>Quickstart</strong>, <strong>Multivariable</strong> <strong>Calculus</strong> -- <strong>Sage</strong>'blue', opacity=0.3)domain = parametric_plot3d([3*u, 4*(3*u)*v,0], (u, 0, 1), (v,0,1), color = 'green', opacity = 0.75)image = parametric_plot3d([3*u, 4*(3*u)*v, f(3*u, 12*u*v)], (u,0, 1), (v, 0,1), color = 'green', opacity = 1.00)surface+domain+imageToggle Advanced ControlsHelp <strong>for</strong> Jmol 3-D viewertest.sagenb.org/home/Carlos_Rodriguez/20/print 9/9