The dual nature of homologous recombination in plants
The dual nature of homologous recombination in plants
The dual nature of homologous recombination in plants
- No tags were found...
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
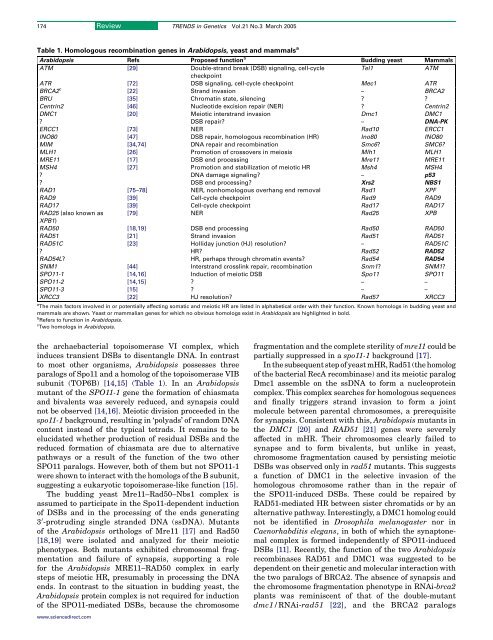
174Review TRENDS <strong>in</strong> Genetics Vol.21 No.3 March 2005Table 1. Homologous <strong>recomb<strong>in</strong>ation</strong> genes <strong>in</strong> Arabidopsis, yeast and mammals aArabidopsis Refs Proposed function b Budd<strong>in</strong>g yeast MammalsATM [29] Double-strand break (DSB) signal<strong>in</strong>g, cell-cycleTel1ATMcheckpo<strong>in</strong>tATR [72] DSB signal<strong>in</strong>g, cell-cycle checkpo<strong>in</strong>t Mec1 ATRBRCA2 c [22] Strand <strong>in</strong>vasion – BRCA2BRU [35] Chromat<strong>in</strong> state, silenc<strong>in</strong>g ? ?Centr<strong>in</strong>2 [46] Nucleotide excision repair (NER) ? Centr<strong>in</strong>2DMC1 [20] Meiotic <strong>in</strong>terstrand <strong>in</strong>vasion Dmc1 DMC1? DSB repair? – DNA-PKERCC1 [73] NER Rad10 ERCC1INO80 [47] DSB repair, <strong>homologous</strong> <strong>recomb<strong>in</strong>ation</strong> (HR) Ino80 INO80MIM [34,74] DNA repair and <strong>recomb<strong>in</strong>ation</strong> Smc6? SMC6?MLH1 [26] Promotion <strong>of</strong> crossovers <strong>in</strong> meiosis Mlh1 MLH1MRE11 [17] DSB end process<strong>in</strong>g Mre11 MRE11MSH4 [27] Promotion and stabilization <strong>of</strong> meiotic HR Msh4 MSH4? DNA damage signal<strong>in</strong>g? – p53? DSB end process<strong>in</strong>g? Xrs2 NBS1RAD1 [75–78] NER, non<strong>homologous</strong> overhang end removal Rad1 XPFRAD9 [39] Cell-cycle checkpo<strong>in</strong>t Rad9 RAD9RAD17 [39] Cell-cycle checkpo<strong>in</strong>t Rad17 RAD17RAD25 (also known as [79] NER Rad25 XPBXPB1)RAD50 [18,19] DSB end process<strong>in</strong>g Rad50 RAD50RAD51 [21] Strand <strong>in</strong>vasion Rad51 RAD51RAD51C [23] Holliday junction (HJ) resolution? – RAD51C? HR? Rad52 RAD52RAD54L? HR, perhaps through chromat<strong>in</strong> events? Rad54 RAD54SNM1 [44] Interstrand crossl<strong>in</strong>k repair, <strong>recomb<strong>in</strong>ation</strong> Snm1? SNM1?SPO11-1 [14,16] Induction <strong>of</strong> meiotic DSB Spo11 SPO11SPO11-2 [14,15] ? – –SPO11-3 [15] ? – –XRCC3 [22] HJ resolution? Rad57 XRCC3a <strong>The</strong> ma<strong>in</strong> factors <strong>in</strong>volved <strong>in</strong> or potentially affect<strong>in</strong>g somatic and meiotic HR are listed <strong>in</strong> alphabetical order with their function. Known homologs <strong>in</strong> budd<strong>in</strong>g yeast andmammals are shown. Yeast or mammalian genes for which no obvious homologs exist <strong>in</strong> Arabidopsis are highlighted <strong>in</strong> bold.b Refers to function <strong>in</strong> Arabidopsis.c Two homologs <strong>in</strong> Arabidopsis.the archaebacterial topoisomerase VI complex, which<strong>in</strong>duces transient DSBs to disentangle DNA. In contrastto most other organisms, Arabidopsis possesses threeparalogs <strong>of</strong> Spo11 and a homolog <strong>of</strong> the topoisomerase VIBsubunit (TOP6B) [14,15] (Table 1). In an Arabidopsismutant <strong>of</strong> the SPO11-1 gene the formation <strong>of</strong> chiasmataand bivalents was severely reduced, and synapsis couldnot be observed [14,16]. Meiotic division proceeded <strong>in</strong> thespo11-1 background, result<strong>in</strong>g <strong>in</strong> ‘polyads’ <strong>of</strong> random DNAcontent <strong>in</strong>stead <strong>of</strong> the typical tetrads. It rema<strong>in</strong>s to beelucidated whether production <strong>of</strong> resi<strong>dual</strong> DSBs and thereduced formation <strong>of</strong> chiasmata are due to alternativepathways or a result <strong>of</strong> the function <strong>of</strong> the two otherSPO11 paralogs. However, both <strong>of</strong> them but not SPO11-1were shown to <strong>in</strong>teract with the homologs <strong>of</strong> the B subunit,suggest<strong>in</strong>g a eukaryotic topoisomerase-like function [15].<strong>The</strong> budd<strong>in</strong>g yeast Mre11–Rad50–Nbs1 complex isassumed to participate <strong>in</strong> the Spo11-dependent <strong>in</strong>duction<strong>of</strong> DSBs and <strong>in</strong> the process<strong>in</strong>g <strong>of</strong> the ends generat<strong>in</strong>g3 0 -protrud<strong>in</strong>g s<strong>in</strong>gle stranded DNA (ssDNA). Mutants<strong>of</strong> the Arabidopsis orthologs <strong>of</strong> Mre11 [17] and Rad50[18,19] were isolated and analyzed for their meioticphenotypes. Both mutants exhibited chromosomal fragmentationand failure <strong>of</strong> synapsis, support<strong>in</strong>g a rolefor the Arabidopsis MRE11–RAD50 complex <strong>in</strong> earlysteps <strong>of</strong> meiotic HR, presumably <strong>in</strong> process<strong>in</strong>g the DNAends. In contrast to the situation <strong>in</strong> budd<strong>in</strong>g yeast, theArabidopsis prote<strong>in</strong> complex is not required for <strong>in</strong>duction<strong>of</strong> the SPO11-mediated DSBs, because the chromosomefragmentation and the complete sterility <strong>of</strong> mre11 could bepartially suppressed <strong>in</strong> a spo11-1 background [17].In the subsequent step <strong>of</strong> yeast mHR, Rad51 (the homolog<strong>of</strong> the bacterial RecA recomb<strong>in</strong>ase) and its meiotic paralogDmc1 assemble on the ssDNA to form a nucleoprote<strong>in</strong>complex. This complex searches for <strong>homologous</strong> sequencesand f<strong>in</strong>ally triggers strand <strong>in</strong>vasion to form a jo<strong>in</strong>tmolecule between parental chromosomes, a prerequisitefor synapsis. Consistent with this, Arabidopsis mutants <strong>in</strong>the DMC1 [20] and RAD51 [21] genes were severelyaffected <strong>in</strong> mHR. <strong>The</strong>ir chromosomes clearly failed tosynapse and to form bivalents, but unlike <strong>in</strong> yeast,chromosome fragmentation caused by persist<strong>in</strong>g meioticDSBs was observed only <strong>in</strong> rad51 mutants. This suggestsa function <strong>of</strong> DMC1 <strong>in</strong> the selective <strong>in</strong>vasion <strong>of</strong> the<strong>homologous</strong> chromosome rather than <strong>in</strong> the repair <strong>of</strong>the SPO11-<strong>in</strong>duced DSBs. <strong>The</strong>se could be repaired byRAD51-mediated HR between sister chromatids or by analternative pathway. Interest<strong>in</strong>gly, a DMC1 homolog couldnot be identified <strong>in</strong> Drosophila melanogaster nor <strong>in</strong>Caenorhabditis elegans, <strong>in</strong> both <strong>of</strong> which the synaptonemalcomplex is formed <strong>in</strong>dependently <strong>of</strong> SPO11-<strong>in</strong>ducedDSBs [11]. Recently, the function <strong>of</strong> the two Arabidopsisrecomb<strong>in</strong>ases RAD51 and DMC1 was suggested to bedependent on their genetic and molecular <strong>in</strong>teraction withthe two paralogs <strong>of</strong> BRCA2. <strong>The</strong> absence <strong>of</strong> synapsis andthe chromosome fragmentation phenotype <strong>in</strong> RNAi-brca2<strong>plants</strong> was rem<strong>in</strong>iscent <strong>of</strong> that <strong>of</strong> the double-mutantdmc1/RNAi-rad51 [22], and the BRCA2 paralogswww.sciencedirect.com