Computational Analysis and Paleogenomics of Interspersed ...
Computational Analysis and Paleogenomics of Interspersed ...
Computational Analysis and Paleogenomics of Interspersed ...
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
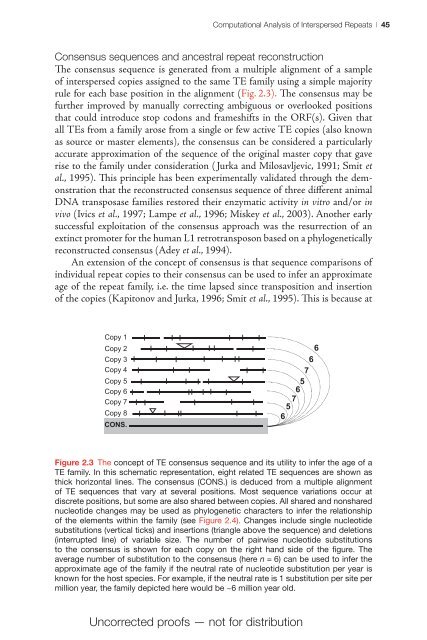
<strong>Computational</strong> <strong>Analysis</strong> <strong>of</strong> <strong>Interspersed</strong> Repeats | 45Consensus sequences <strong>and</strong> ancestral repeat reconstructionThe consensus sequence is generated from a multiple alignment <strong>of</strong> a sample<strong>of</strong> interspersed copies assigned to the same TE family using a simple majorityrule for each base position in the alignment (Fig. 2.3). The consensus may befurther improved by manually correcting ambiguous or overlooked positionsthat could introduce stop codons <strong>and</strong> frameshifts in the ORF(s). Given thatall TEs from a family arose from a single or few active TE copies (also knownas source or master elements), the consensus can be considered a particularlyaccurate approximation <strong>of</strong> the sequence <strong>of</strong> the original master copy that gaverise to the family under consideration ( Jurka <strong>and</strong> Milosavljevic, 1991; Smit etal., 1995). This principle has been experimentally validated through the demonstrationthat the reconstructed consensus sequence <strong>of</strong> three different animalDNA transposase families restored their enzymatic activity in vitro <strong>and</strong>/or invivo (Ivics et al., 1997; Lampe et al., 1996; Miskey et al., 2003). Another earlysuccessful exploitation <strong>of</strong> the consensus approach was the resurrection <strong>of</strong> anextinct promoter for the human L1 retrotransposon based on a phylogeneticallyreconstructed consensus (Adey et al., 1994).An extension <strong>of</strong> the concept <strong>of</strong> consensus is that sequence comparisons <strong>of</strong>individual repeat copies to their consensus can be used to infer an approximateage <strong>of</strong> the repeat family, i.e. the time lapsed since transposition <strong>and</strong> insertion<strong>of</strong> the copies (Kapitonov <strong>and</strong> Jurka, 1996; Smit et al., 1995). This is because atCopy 1Copy 2Copy 3Copy 4Copy 5Copy 6Copy 7Copy 8CONS.66756756Figure 2.3 The concept <strong>of</strong> TE consensus sequence <strong>and</strong> its utility to infer the age <strong>of</strong> aTE family. In this schematic representation, eight related TE sequences are shown asthick horizontal lines. The consensus (CONS.) is deduced from a multiple alignment<strong>of</strong> TE sequences that vary at several Figure positions. 3 Most sequence variations occur atdiscrete positions, but some are also shared between copies. All shared <strong>and</strong> nonsharednucleotide changes may be used as phylogenetic characters to infer the relationship<strong>of</strong> the elements within the family (see Figure 2.4). Changes include single nucleotidesubstitutions (vertical ticks) <strong>and</strong> insertions (triangle above the sequence) <strong>and</strong> deletions(interrupted line) <strong>of</strong> variable size. The number <strong>of</strong> pairwise nucleotide substitutionsto the consensus is shown for each copy on the right h<strong>and</strong> side <strong>of</strong> the figure. Theaverage number <strong>of</strong> substitution to the consensus (here n = 6) can be used to infer theapproximate age <strong>of</strong> the family if the neutral rate <strong>of</strong> nucleotide substitution per year isknown for the host species. For example, if the neutral rate is 1 substitution per site permillion year, the family depicted here would be ~6 million year old.Uncorrected pro<strong>of</strong>s — not for distribution