Phylogeny and classification of the Digenea (Platyhelminthes ...
Phylogeny and classification of the Digenea (Platyhelminthes ...
Phylogeny and classification of the Digenea (Platyhelminthes ...
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
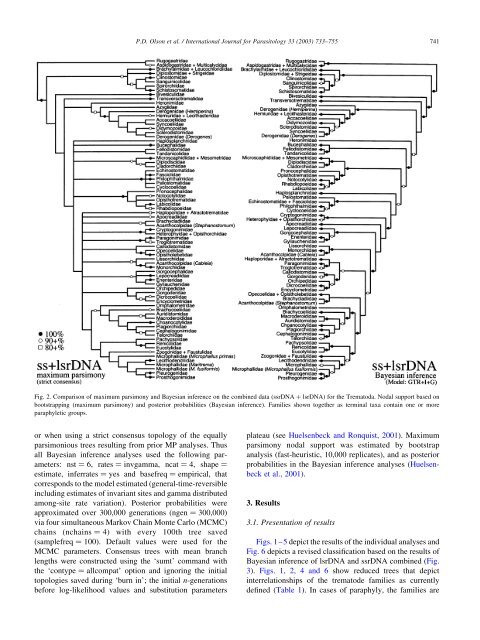
P.D. Olson et al. / International Journal for Parasitology 33 (2003) 733–755 741Fig. 2. Comparison <strong>of</strong> maximum parsimony <strong>and</strong> Bayesian inference on <strong>the</strong> combined data (ssrDNA þ lsrDNA) for <strong>the</strong> Trematoda. Nodal support based onbootstrapping (maximum parsimony) <strong>and</strong> posterior probabilities (Bayesian inference). Families shown toge<strong>the</strong>r as terminal taxa contain one or moreparaphyletic groups.or when using a strict consensus topology <strong>of</strong> <strong>the</strong> equallyparsimonious trees resulting from prior MP analyses. Thusall Bayesian inference analyses used <strong>the</strong> following parameters:nst ¼ 6, rates ¼ invgamma, ncat ¼ 4, shape ¼estimate, inferrates ¼ yes <strong>and</strong> basefreq ¼ empirical, thatcorresponds to <strong>the</strong> model estimated (general-time-reversibleincluding estimates <strong>of</strong> invariant sites <strong>and</strong> gamma distributedamong-site rate variation). Posterior probabilities wereapproximated over 300,000 generations (ngen ¼ 300,000)via four simultaneous Markov Chain Monte Carlo (MCMC)chains (nchains ¼ 4) with every 100th tree saved(samplefreq ¼ 100). Default values were used for <strong>the</strong>MCMC parameters. Consensus trees with mean branchlengths were constructed using <strong>the</strong> ‘sumt’ comm<strong>and</strong> with<strong>the</strong> ‘contype ¼ allcompat’ option <strong>and</strong> ignoring <strong>the</strong> initialtopologies saved during ‘burn in’; <strong>the</strong> initial n-generationsbefore log-likelihood values <strong>and</strong> substitution parametersplateau (see Huelsenbeck <strong>and</strong> Ronquist, 2001). Maximumparsimony nodal support was estimated by bootstrapanalysis (fast-heuristic, 10,000 replicates), <strong>and</strong> as posteriorprobabilities in <strong>the</strong> Bayesian inference analyses (Huelsenbecket al., 2001).3. Results3.1. Presentation <strong>of</strong> resultsFigs. 1–5 depict <strong>the</strong> results <strong>of</strong> <strong>the</strong> individual analyses <strong>and</strong>Fig. 6 depicts a revised <strong>classification</strong> based on <strong>the</strong> results <strong>of</strong>Bayesian inference <strong>of</strong> lsrDNA <strong>and</strong> ssrDNA combined (Fig.3). Figs. 1, 2, 4 <strong>and</strong> 6 show reduced trees that depictinterrelationships <strong>of</strong> <strong>the</strong> trematode families as currentlydefined (Table 1). In cases <strong>of</strong> paraphyly, <strong>the</strong> families are