Raman spectroscopy of biological pigments Solid mixed matrices in MALDI/TOF-MS
1VpTi1r
1VpTi1r
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
VOL. 28 NO. 1 (2016)<br />
TONY DAVIES COLUMN<br />
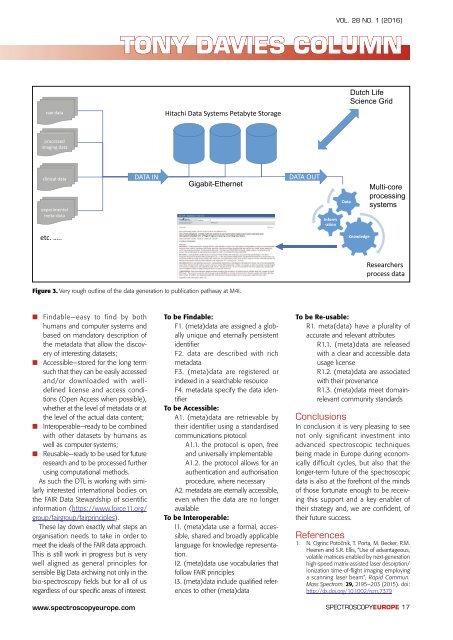
raw data<br />
Hitachi Data Systems Petabyte Storage<br />
Dutch Life<br />
Science Grid<br />
processed<br />
imag<strong>in</strong>g data<br />
cl<strong>in</strong>ical data<br />
experimental<br />
meta‐data<br />
DATA IN<br />
Gigabit-Ethernet<br />
DATA OUT<br />
Inform<br />
‐ation<br />
Data<br />
Multi-core<br />
process<strong>in</strong>g<br />
systems<br />
etc. …..<br />
Knowledge<br />
Researchers<br />
process data<br />
Figure 3. Very rough outl<strong>in</strong>e <strong>of</strong> the data generation to publication pathway at M4I.<br />
■■<br />
■■<br />
■■<br />
■■<br />
F<strong>in</strong>dable—easy to f<strong>in</strong>d by both<br />
humans and computer systems and<br />
based on mandatory description <strong>of</strong><br />
the metadata that allow the discovery<br />
<strong>of</strong> <strong>in</strong>terest<strong>in</strong>g datasets;<br />
Accessible—stored for the long term<br />
such that they can be easily accessed<br />
and/or downloaded with welldef<strong>in</strong>ed<br />
license and access conditions<br />
(Open Access when possible),<br />
whether at the level <strong>of</strong> metadata or at<br />
the level <strong>of</strong> the actual data content;<br />
Interoperable—ready to be comb<strong>in</strong>ed<br />
with other datasets by humans as<br />
well as computer systems;<br />
Reusable—ready to be used for future<br />
research and to be processed further<br />
us<strong>in</strong>g computational methods.<br />
As such the DTL is work<strong>in</strong>g with similarly<br />
<strong>in</strong>terested <strong>in</strong>ternational bodies on<br />
the FAIR Data Stewardship <strong>of</strong> scientific<br />
<strong>in</strong>formation (https://www.force11.org/<br />
group/fairgroup/fairpr<strong>in</strong>ciples).<br />
These lay down exactly what steps an<br />
organisation needs to take <strong>in</strong> order to<br />
meet the ideals <strong>of</strong> the FAIR data approach.<br />
This is still work <strong>in</strong> progress but is very<br />
well aligned as general pr<strong>in</strong>ciples for<br />
sensible Big Data archiv<strong>in</strong>g not only <strong>in</strong> the<br />
bio-<strong>spectroscopy</strong> fields but for all <strong>of</strong> us<br />
regardless <strong>of</strong> our specific areas <strong>of</strong> <strong>in</strong>terest.<br />
www.<strong>spectroscopy</strong>europe.com<br />
To be F<strong>in</strong>dable:<br />
F1. (meta)data are assigned a globally<br />
unique and eternally persistent<br />
identifier<br />
F2. data are described with rich<br />
metadata<br />
F3. (meta)data are registered or<br />
<strong>in</strong>dexed <strong>in</strong> a searchable resource<br />
F4. metadata specify the data identifier<br />
To be Accessible:<br />
A1. (meta)data are retrievable by<br />
their identifier us<strong>in</strong>g a standardised<br />
communications protocol<br />
A1.1. the protocol is open, free<br />
and universally implementable<br />
A1.2. the protocol allows for an<br />
authentication and authorisation<br />
procedure, where necessary<br />
A2. metadata are eternally accessible,<br />
even when the data are no longer<br />
available<br />
To be Interoperable:<br />
I1. (meta)data use a formal, accessible,<br />
shared and broadly applicable<br />
language for knowledge representation.<br />
I2. (meta)data use vocabularies that<br />
follow FAIR pr<strong>in</strong>ciples<br />
I3. (meta)data <strong>in</strong>clude qualified references<br />
to other (meta)data<br />
To be Re-usable:<br />
R1. meta(data) have a plurality <strong>of</strong><br />
accurate and relevant attributes<br />
R1.1. (meta)data are released<br />
with a clear and accessible data<br />
usage license<br />
R1.2. (meta)data are associated<br />
with their provenance<br />
R1.3. (meta)data meet doma<strong>in</strong>relevant<br />
community standards<br />
Conclusions<br />
In conclusion it is very pleas<strong>in</strong>g to see<br />
not only significant <strong>in</strong>vestment <strong>in</strong>to<br />
advanced spectroscopic techniques<br />
be<strong>in</strong>g made <strong>in</strong> Europe dur<strong>in</strong>g economically<br />
difficult cycles, but also that the<br />
longer-term future <strong>of</strong> the spectroscopic<br />
data is also at the forefront <strong>of</strong> the m<strong>in</strong>ds<br />
<strong>of</strong> those fortunate enough to be receiv<strong>in</strong>g<br />
this support and a key enabler <strong>of</strong><br />
their strategy and, we are confident, <strong>of</strong><br />
their future success.<br />
References<br />
1. N. Ogr<strong>in</strong>c Potočnik, T. Porta, M. Becker, R.M.<br />
Heeren and S.R. Ellis, “Use <strong>of</strong> advantageous,<br />
volatile <strong>matrices</strong> enabled by next-generation<br />
high-speed matrix-assisted laser desorption/<br />
ionization time-<strong>of</strong>-flight imag<strong>in</strong>g employ<strong>in</strong>g<br />
a scann<strong>in</strong>g laser beam”, Rapid Commun.<br />
Mass Spectrom. 29, 2195–203 (2015). doi:<br />
http://dx.doi.org/10.1002/rcm.7379<br />
SPECTROSCOPYEUROPE 17