Phylogeny and molecular evolution of green algae - Phycology ...
Phylogeny and molecular evolution of green algae - Phycology ...
Phylogeny and molecular evolution of green algae - Phycology ...
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
22 CHAPTER 2<br />
underst<strong>and</strong>ing this diversification is to resolve the phylogenetic relationships among the UTC classes<br />
<strong>and</strong> the ulvophycean orders. The fact that previous <strong>molecular</strong> phylogenetic analyses have not yielded<br />
a satisfactory resolution <strong>of</strong> these taxa can be attributed at least in part to the difficulty in obtaining a<br />
good combination <strong>of</strong> taxon <strong>and</strong> gene sampling. Single-gene analyses with good taxon sampling have<br />
suffered from poor resolving power due to the low number <strong>of</strong> characters (Watanabe <strong>and</strong> Nakayama<br />
2007). Conversely, studies based on longer alignments from complete organelle genomes have<br />
suffered from sparse <strong>and</strong> uneven taxon sampling (Pombert et al. 2004, Pombert et al. 2005), which<br />
can lead to systematic error in phylogenetic analysis (Brinkmann et al. 2005, Philippe et al. 2005).<br />
Our goal in this study is to infer ancient relationships within the Chlorophyta, more specifically the<br />
divergence <strong>of</strong> UTC classes <strong>and</strong> ulvophycean orders. Our approach consists <strong>of</strong> phylogenetic analysis <strong>of</strong><br />
a dataset that <strong>of</strong>fers a good balance between taxon <strong>and</strong> gene sampling. Our alignment consists <strong>of</strong><br />
seven single-copy nuclear markers, SSU nrDNA <strong>and</strong> two plastid genes for 43 taxa representing the<br />
major lineages <strong>of</strong> the Viridiplantae. Phylogenetic analyses are carried out with model-based<br />
techniques, paying careful attention to the selection <strong>of</strong> suitable partitioning strategies <strong>and</strong> models <strong>of</strong><br />
sequence <strong>evolution</strong>. Noise-reduction techniques targeted at improving signal in the epoch <strong>of</strong> interest<br />
are applied.<br />
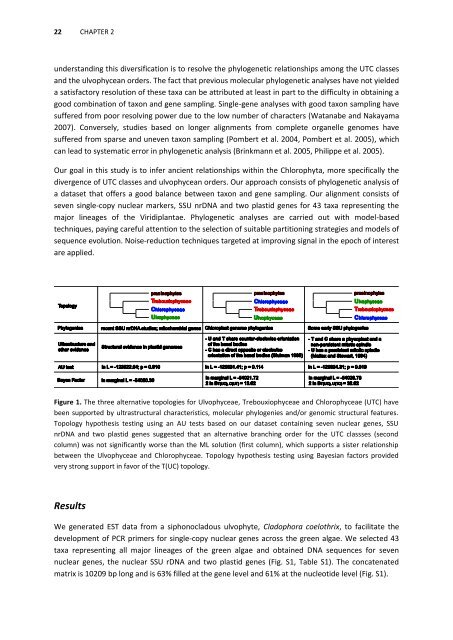
Figure 1. The three alternative topologies for Ulvophyceae, Trebouxiophyceae <strong>and</strong> Chlorophyceae (UTC) have<br />
been supported by ultrastructural characteristics, <strong>molecular</strong> phylogenies <strong>and</strong>/or genomic structural features.<br />
Topology hypothesis testing using an AU tests based on our dataset containing seven nuclear genes, SSU<br />
nrDNA <strong>and</strong> two plastid genes suggested that an alternative branching order for the UTC classses (second<br />
column) was not significantly worse than the ML solution (first column), which supports a sister relationship<br />
between the Ulvophyceae <strong>and</strong> Chlorophyceae. Topology hypothesis testing using Bayesian factors provided<br />
very strong support in favor <strong>of</strong> the T(UC) topology.<br />
Results<br />
We generated EST data from a siphonocladous ulvophyte, Cladophora coelothrix, to facilitate the<br />
development <strong>of</strong> PCR primers for single-copy nuclear genes across the <strong>green</strong> <strong>algae</strong>. We selected 43<br />
taxa representing all major lineages <strong>of</strong> the <strong>green</strong> <strong>algae</strong> <strong>and</strong> obtained DNA sequences for seven<br />
nuclear genes, the nuclear SSU rDNA <strong>and</strong> two plastid genes (Fig. S1, Table S1). The concatenated<br />
matrix is 10209 bp long <strong>and</strong> is 63% filled at the gene level <strong>and</strong> 61% at the nucleotide level (Fig. S1).