Intraspecific Variation in Viola suavis in Europe ... - Annals of Botany
Intraspecific Variation in Viola suavis in Europe ... - Annals of Botany
Intraspecific Variation in Viola suavis in Europe ... - Annals of Botany
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
450<br />
A<br />
90<br />
B<br />
81<br />
Estimation <strong>of</strong> pollen fertility<br />
0·001 changes<br />
To test the assumed hybrid orig<strong>in</strong> <strong>of</strong> the white-flowered<br />
morphotype(s), all plants <strong>in</strong>cluded <strong>in</strong> morphometric analyses<br />
were also exam<strong>in</strong>ed for pollen viability. Anthers<br />
were removed from a s<strong>in</strong>gle flower or a flower bud per <strong>in</strong>dividual<br />
and chopped <strong>in</strong> a drop <strong>of</strong> aceto-carm<strong>in</strong>e jelly<br />
(Radford et al., 1974) on a microscope slide to release<br />
pollen gra<strong>in</strong>s. One hundred pollen gra<strong>in</strong>s per <strong>in</strong>dividual<br />
were observed. The numbers <strong>of</strong> unviable (i.e. unsta<strong>in</strong>ed<br />
and usually shrunken) and viable (well-sta<strong>in</strong>ed and <strong>of</strong><br />
regular shape) gra<strong>in</strong>s were recorded. Pollen quality was<br />
expressed as the percentage <strong>of</strong> viable gra<strong>in</strong>s.<br />
RESULTS<br />
Karyological analyses<br />
Data on chromosome number and DNA ploidy level are<br />
presented <strong>in</strong> Appendix. All populations <strong>of</strong> V. alba,<br />
V. coll<strong>in</strong>a, V. hirta and V. odorata were found to be tetraploid<br />
with 2n ¼ 20, while all populations <strong>of</strong> V. <strong>suavis</strong><br />
(both morphotypes) were octoploid with 2n ¼ 40. These<br />
data are consistent with previous reports (cf. Mered’a<br />
et al., 2006; Hodálová et al., 2008).<br />
Molecular analyses<br />
ITS sequences. The ITS matrix exclud<strong>in</strong>g the outgroup consisted<br />
<strong>of</strong> 38 sequences and 612 aligned positions; 37 sites<br />
were variable; 34 <strong>of</strong> them were parsimony <strong>in</strong>formative.<br />
A few 1-bp or 2-bp <strong>in</strong>dels were identified, which were<br />
not coded separately but treated as miss<strong>in</strong>g data. Only a<br />
few sequences conta<strong>in</strong>ed apparent <strong>in</strong>tra-<strong>in</strong>dividual s<strong>in</strong>glesite<br />
polymorphisms (overlapp<strong>in</strong>g peaks <strong>in</strong> electropherograms;<br />
up to two positions per sequence). The parsimony<br />
analysis (figure not shown) resulted <strong>in</strong> a strict consensus<br />
tree (2288 most parsimonious trees; L ¼ 50 steps,<br />
Mered’a et al. — <strong>Intraspecific</strong> <strong>Variation</strong> <strong>in</strong> <strong>Viola</strong> <strong>suavis</strong><br />
71<br />
100<br />
100<br />
AMB 149a<br />
AMB 149b<br />
95 AMB 151a<br />
59<br />
AMB 151c<br />
AMB 151b<br />
AMB 149c<br />
65<br />
62 AMB 21a<br />
AMB 21c<br />
AMB 21b<br />
76<br />
ambigua<br />
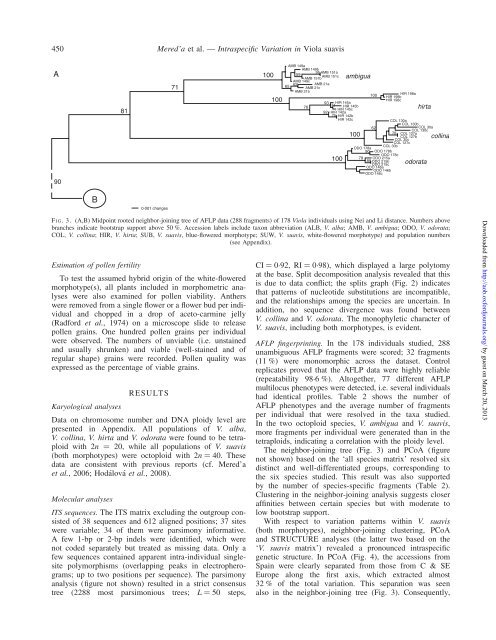
CI ¼ 0.92, RI ¼ 0.98), which displayed a large polytomy<br />
at the base. Split decomposition analysis revealed that this<br />
is due to data conflict; the splits graph (Fig. 2) <strong>in</strong>dicates<br />
that patterns <strong>of</strong> nucleotide substitutions are <strong>in</strong>compatible,<br />
and the relationships among the species are uncerta<strong>in</strong>. In<br />
addition, no sequence divergence was found between<br />
V. coll<strong>in</strong>a and V. odorata. The monophyletic character <strong>of</strong><br />
V. <strong>suavis</strong>, <strong>in</strong>clud<strong>in</strong>g both morphotypes, is evident.<br />
AFLP f<strong>in</strong>gerpr<strong>in</strong>t<strong>in</strong>g. In the 178 <strong>in</strong>dividuals studied, 288<br />
unambiguous AFLP fragments were scored; 32 fragments<br />
(11 %) were monomorphic across the dataset. Control<br />
replicates proved that the AFLP data were highly reliable<br />
(repeatability 98.6 %). Altogether, 77 different AFLP<br />
multilocus phenotypes were detected, i.e. several <strong>in</strong>dividuals<br />
had identical pr<strong>of</strong>iles. Table 2 shows the number <strong>of</strong><br />
AFLP phenotypes and the average number <strong>of</strong> fragments<br />
per <strong>in</strong>dividual that were resolved <strong>in</strong> the taxa studied.<br />
In the two octoploid species, V. ambigua and V. <strong>suavis</strong>,<br />
more fragments per <strong>in</strong>dividual were generated than <strong>in</strong> the<br />
tetraploids, <strong>in</strong>dicat<strong>in</strong>g a correlation with the ploidy level.<br />
The neighbor-jo<strong>in</strong><strong>in</strong>g tree (Fig. 3) and PCoA (figure<br />
not shown) based on the ‘all species matrix’ resolved six<br />
dist<strong>in</strong>ct and well-differentiated groups, correspond<strong>in</strong>g to<br />
the six species studied. This result was also supported<br />
by the number <strong>of</strong> species-specific fragments (Table 2).<br />
Cluster<strong>in</strong>g <strong>in</strong> the neighbor-jo<strong>in</strong><strong>in</strong>g analysis suggests closer<br />
aff<strong>in</strong>ities between certa<strong>in</strong> species but with moderate to<br />
low bootstrap support.<br />
With respect to variation patterns with<strong>in</strong> V. <strong>suavis</strong><br />
(both morphotypes), neighbor-jo<strong>in</strong><strong>in</strong>g cluster<strong>in</strong>g, PCoA<br />
and STRUCTURE analyses (the latter two based on the<br />
‘V. <strong>suavis</strong> matrix’) revealed a pronounced <strong>in</strong>traspecific<br />
genetic structure. In PCoA (Fig. 4), the accessions from<br />
Spa<strong>in</strong> were clearly separated from those from C & SE<br />
<strong>Europe</strong> along the first axis, which extracted almost<br />
32 % <strong>of</strong> the total variation. This separation was seen<br />
also <strong>in</strong> the neighbor-jo<strong>in</strong><strong>in</strong>g tree (Fig. 3). Consequently,<br />
100<br />
HIR 198a<br />
HIR 198b<br />
HIR 198c<br />
93 HIR 145a<br />
71 HIR 145b<br />
HIR 145c<br />
hirta<br />
92 HIR 142a<br />
75 HIR 142b<br />
HIR 142c<br />
COL 130a<br />
COL 130bCOL<br />
62<br />
30a<br />
COL 130c<br />
100<br />
78 COL 127a<br />
COL 127b<br />
COL 30c<br />
COL 127c<br />
COL 30b<br />
ODO 178a<br />
90 ODO 178b<br />
ODO 178c<br />
100 79 ODO 215a<br />
89 ODO 215b<br />
ODO 215c odorata<br />
ODO 146a<br />
ODO 146b<br />
ODO 146c<br />
F IG. 3. (A,B) Midpo<strong>in</strong>t rooted neighbor-jo<strong>in</strong><strong>in</strong>g tree <strong>of</strong> AFLP data (288 fragments) <strong>of</strong> 178 <strong>Viola</strong> <strong>in</strong>dividuals us<strong>in</strong>g Nei and Li distance. Numbers above<br />
branches <strong>in</strong>dicate bootstrap support above 50 %. Accession labels <strong>in</strong>clude taxon abbreviation (ALB, V. alba; AMB, V. ambigua; ODO, V. odorata;<br />
COL, V. coll<strong>in</strong>a; HIR, V. hirta; SUB, V. <strong>suavis</strong>, blue-flowered morphotype; SUW, V. <strong>suavis</strong>, white-flowered morphotype) and population numbers<br />
(see Appendix).<br />
coll<strong>in</strong>a<br />
Downloaded from<br />
http://aob.oxfordjournals.org/<br />
by guest on March 20, 2013